|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bS,5aR,9aR,9bS,11aS)-N-[1-(4-chlorophenyl)cyclopentyl]-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,
(1S,3aS,3bS,5aR,9aR,9bS,11aS)-N-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.58 |
12.11 |
-13.07 |
2 |
4 |
0 |
58 |
497.123 |
3 |
↓
|
|
|
|
Analogs
-
3799355
-
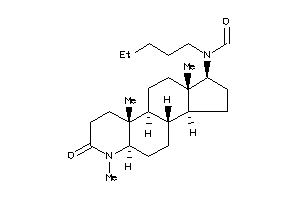
-
3799357
-
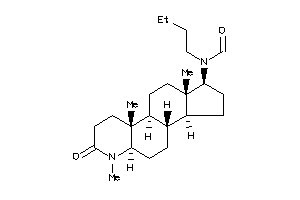
-
3799359
-
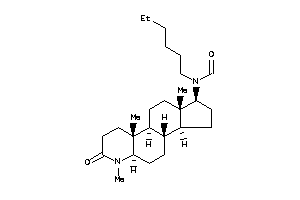
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecahydro-
N-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
7.81 |
-13.7 |
1 |
4 |
0 |
49 |
346.515 |
1 |
↓
|
|
|
|
Analogs
-
3799355
-

-
3799357
-

-
3799359
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[(1R,3aR,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecahydro-
N-[(1R,3aR,3bR,5aR,9aR,9bR,11aR)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
8.08 |
-12.41 |
1 |
4 |
0 |
49 |
346.515 |
1 |
↓
|
|
|
|
Analogs
-
36152986
-
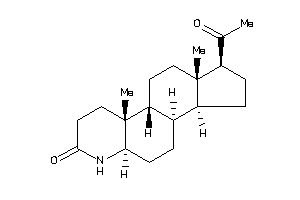
-
36152989
-
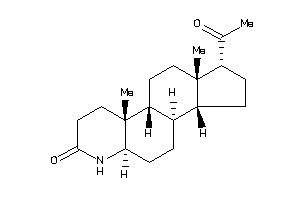
-
36152993
-
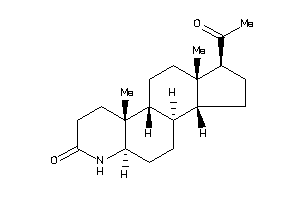
-
5275340
-
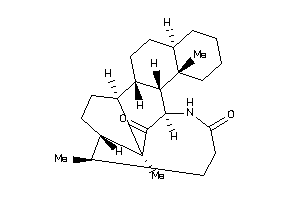
-
5275341
-
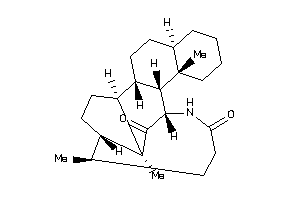
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-1-acetyl-9a,11a-dimethyl-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-tetradecahy
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-1-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
7.64 |
-9.73 |
1 |
3 |
0 |
46 |
317.473 |
1 |
↓
|
|
|
|
Analogs
-
34212569
-
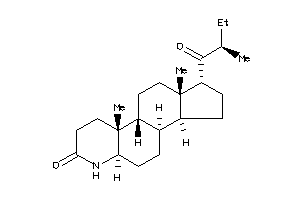
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-1-acetyl-9a,11a-dimethyl-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-tetradecahy
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-1-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
7.59 |
-10.62 |
1 |
3 |
0 |
46 |
317.473 |
1 |
↓
|
|
|
|
Analogs
-
34212569
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-1-acetyl-9a,11a-dimethyl-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-tetradecahy
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-1-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
7.04 |
-8.5 |
1 |
3 |
0 |
46 |
317.473 |
1 |
↓
|
|
|
|
Analogs
-
34212569
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-1-acetyl-9a,11a-dimethyl-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-tetradecahy
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-1-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
7.53 |
-10.81 |
1 |
3 |
0 |
46 |
317.473 |
1 |
↓
|
|
|
|
Analogs
-
36153001
-
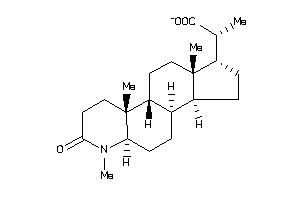
-
36153005
-
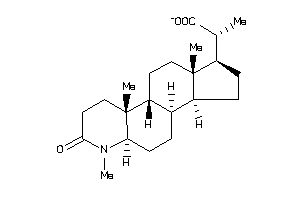
-
6030241
-
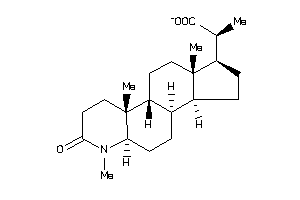
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-2-[(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(2S)-2-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
S5A2-2-E |
Steroid 5-alpha-reductase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
10.53 |
-53.96 |
0 |
4 |
-1 |
60 |
360.518 |
2 |
↓
|
|
|
|
Analogs
-
36153005
-

-
36152997
-
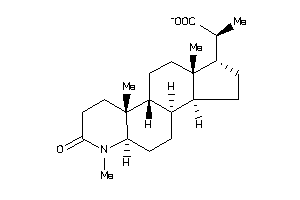
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-2-[(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(2R)-2-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
S5A2-2-E |
Steroid 5-alpha-reductase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
10.22 |
-52.11 |
0 |
4 |
-1 |
60 |
360.518 |
2 |
↓
|
|
|
|
Analogs
-
36152997
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-2-[(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(2R)-2-[(1R,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
S5A2-2-E |
Steroid 5-alpha-reductase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
10.5 |
-49.69 |
0 |
4 |
-1 |
60 |
360.518 |
2 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9S,9aR,9bR,11aS)-N,N-diethyl-6,9,9a,11a-tetramethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10
(1R,3aS,3bR,5aR,9S,9aR,9bR,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
11.18 |
-11.59 |
0 |
4 |
0 |
41 |
402.623 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9S,9aR,9bR,11aS)-N,N-diethyl-6,9,9a,11a-tetramethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10
(1S,3aS,3bR,5aR,9S,9aR,9bR,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
11.04 |
-12.64 |
0 |
4 |
0 |
41 |
402.623 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9S,9aR,9bR,11aS)-N,N-diethyl-6,9,9a,11a-tetramethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10
(1R,3aR,3bR,5aR,9S,9aR,9bR,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
10.23 |
-6.65 |
0 |
4 |
0 |
41 |
402.623 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9S,9aR,9bR,11aS)-N,N-diethyl-6,9,9a,11a-tetramethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10
(1S,3aR,3bR,5aR,9S,9aR,9bR,11aS)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
10.77 |
-12.47 |
0 |
4 |
0 |
41 |
402.623 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N-octadecyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.52 |
20.38 |
-12.79 |
1 |
4 |
0 |
49 |
584.974 |
18 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N-octadecyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.52 |
20.57 |
-11.19 |
1 |
4 |
0 |
49 |
584.974 |
18 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N-octadecyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.52 |
20.47 |
-8.96 |
1 |
4 |
0 |
49 |
584.974 |
18 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N-octadecyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.52 |
20.28 |
-12.52 |
1 |
4 |
0 |
49 |
584.974 |
18 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aS,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1R,3aS,3bR,5aS,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
12.2 |
-11.93 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aS,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1S,3aS,3bR,5aS,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.95 |
-11.15 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aS,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1R,3aR,3bR,5aS,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.11 |
-6.85 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aS,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1S,3aR,3bR,5aS,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.88 |
-11.61 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N,N-dioctyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.18 |
20.24 |
-11.51 |
0 |
4 |
0 |
41 |
556.92 |
15 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N,N-dioctyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.18 |
20.13 |
-12.45 |
0 |
4 |
0 |
41 |
556.92 |
15 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N,N-dioctyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.18 |
19.58 |
-7.27 |
0 |
4 |
0 |
41 |
556.92 |
15 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-N,N-dioctyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dod
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-6,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.18 |
19.85 |
-12.66 |
0 |
4 |
0 |
41 |
556.92 |
15 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-2-[(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(2S)-2-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
12.04 |
-12.71 |
0 |
4 |
0 |
41 |
416.65 |
4 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-2-[(1S,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(2R)-2-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
11.58 |
-11.01 |
0 |
4 |
0 |
41 |
416.65 |
4 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-2-[(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(2S)-2-[(1R,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
11.8 |
-14.04 |
0 |
4 |
0 |
41 |
416.65 |
4 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-2-[(1R,3aS,3bR,5aR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(2R)-2-[(1R,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
11.66 |
-11.84 |
0 |
4 |
0 |
41 |
416.65 |
4 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,1
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
11 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
11 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
10.05 |
-11.69 |
1 |
4 |
0 |
49 |
402.623 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,1
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
11 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
11 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
9.38 |
-5.86 |
1 |
4 |
0 |
49 |
402.623 |
3 |
↓
|
|
|
|
Analogs
-
36153128
-
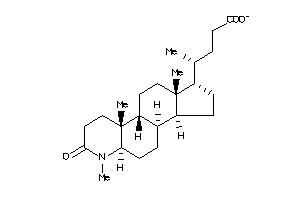
-
36153132
-
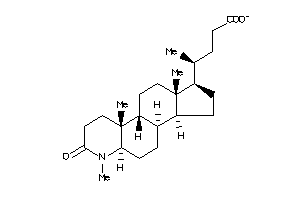
-
36153136
-
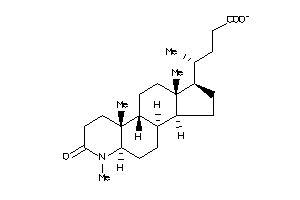
-
40843988
-
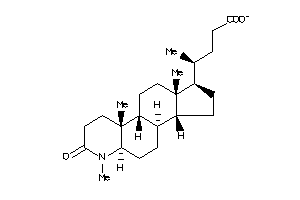
-
40843990
-
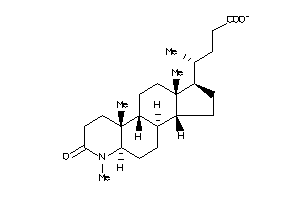
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S)-4-[(1S,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(4S)-4-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-2-E |
Steroid 5-alpha-reductase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
11.6 |
-51.47 |
0 |
4 |
-1 |
60 |
388.572 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
9.63 |
-9.97 |
1 |
4 |
0 |
58 |
389.58 |
4 |
↓
|
|
|
|
Analogs
-
36153132
-

-
36153136
-

-
40843988
-

-
40843990
-

-
3946711
-
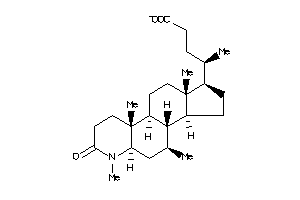
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R)-4-[(1S,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(4R)-4-[(1S,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-2-E |
Steroid 5-alpha-reductase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
11.76 |
-49.89 |
0 |
4 |
-1 |
60 |
388.572 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
9.78 |
-11.34 |
1 |
4 |
0 |
58 |
389.58 |
4 |
↓
|
|
|
|
Analogs
-
36153136
-

-
40843988
-

-
40843990
-

-
3946711
-

-
36153123
-
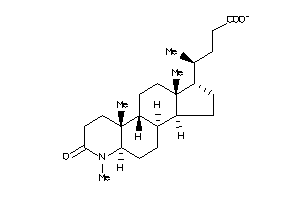
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S)-4-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(4S)-4-[(1R,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-2-E |
Steroid 5-alpha-reductase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
11.87 |
-50.2 |
0 |
4 |
-1 |
60 |
388.572 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
9.89 |
-10.54 |
1 |
4 |
0 |
58 |
389.58 |
4 |
↓
|
|
|
|
Analogs
-
40843988
-

-
40843990
-

-
3946711
-

-
36153123
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R)-4-[(1R,3aS,3bR,5aR,9aR,9bR,11aR)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11-dodecah
(4R)-4-[(1R,3aS,3bR,5aR,9aR,9bR,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-2-E |
Steroid 5-alpha-reductase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
11.89 |
-50.13 |
0 |
4 |
-1 |
60 |
388.572 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
9.92 |
-11.28 |
1 |
4 |
0 |
58 |
389.58 |
4 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.9 |
-11.89 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.61 |
-11.51 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
10.75 |
-7.27 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diisopropyl-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,10,11
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.48 |
-11.73 |
0 |
4 |
0 |
41 |
416.65 |
3 |
↓
|
|
|
|
Analogs
-
36153197
-
-
36153200
-
-
36153205
-
Draw
Identity
99%
90%
80%
70%
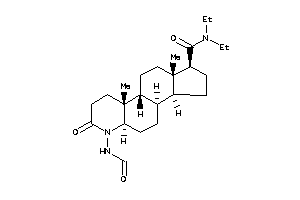
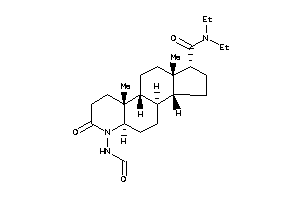
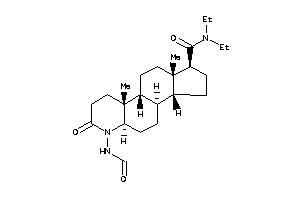
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-6-formamido-9a,11a-dimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
9.55 |
-15.37 |
1 |
6 |
0 |
70 |
417.594 |
4 |
↓
|
|
|
|
Analogs
-
36153200
-

-
36153205
-

-
36153191
-
Draw
Identity
99%
90%
80%
70%
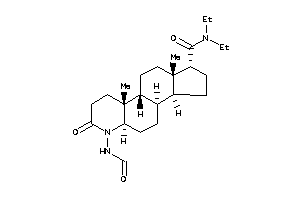
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-6-formamido-9a,11a-dimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
9.44 |
-17.09 |
1 |
6 |
0 |
70 |
417.594 |
4 |
↓
|
|
|
|
Analogs
-
36153205
-

-
36153191
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-6-formamido-9a,11a-dimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
9.17 |
-9.5 |
1 |
6 |
0 |
70 |
417.594 |
4 |
↓
|
|
|
|
Analogs
-
36153191
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-6-formamido-9a,11a-dimethyl-7-oxo-2,3,3a,3b,4,5,5a,8,9,9b,
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
9.36 |
-17.99 |
1 |
6 |
0 |
70 |
417.594 |
4 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-te
(1R,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
9.05 |
-11.9 |
1 |
4 |
0 |
49 |
374.569 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-te
(1S,3aS,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
8.91 |
-12.41 |
1 |
4 |
0 |
49 |
374.569 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-te
(1R,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
8.67 |
-6.26 |
1 |
4 |
0 |
49 |
374.569 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,N-diethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-te
(1S,3aR,3bR,5aR,9aR,9bR,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
8.84 |
-12.37 |
1 |
4 |
0 |
49 |
374.569 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,5aR,9aR,9bS,11aS)-N,N-diethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-te
(1S,3aS,3bR,5aR,9aR,9bS,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
8.84 |
-12.73 |
1 |
4 |
0 |
49 |
374.569 |
3 |
↓
|
|
|
|
Analogs
-
5649661
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,5aR,9aR,9bS,11aS)-N,N-diethyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,5a,6,8,9,9b,10,11-te
(1S,3aR,3bR,5aR,9aR,9bS,11aS)-N,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-2-E |
Steroid 5-alpha-reductase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
8.68 |
-13.25 |
1 |
4 |
0 |
49 |
374.569 |
3 |
↓
|
|