|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
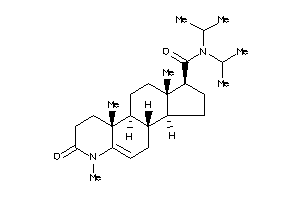
(1S,3aS,3bS,9aR,9bS,11aS)-N-[1-(4-chlorophenyl)cyclopentyl]-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,6,8,
(1S,3aS,3bS,9aR,9bS,11aS)-N-[1-(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.95 |
12.59 |
-13 |
2 |
4 |
0 |
58 |
495.107 |
3 |
↓
|
|
|
|
Analogs
-
37117478
-

-
37117479
-

-
45284557
-
-
34801270
-

-
34801271
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,6,8,9,9b,10,11-dodecah
(1R,3aS,3bR,9aR,9bR,11aS)-N,N-di…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
10.76 |
-11.57 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
-
37117478
-

-
37117479
-

-
45284557
-

-
34801270
-

-
34801271
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aS,3bR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,6,8,9,9b,10,11-dodecah
(1S,3aS,3bR,9aR,9bR,11aS)-N,N-di…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
10.49 |
-10.82 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
-
37117478
-

-
37117479
-

-
45284557
-

-
34801270
-

-
34801271
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,3bR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,6,8,9,9b,10,11-dodecah
(1R,3aR,3bR,9aR,9bR,11aS)-N,N-di…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
9.78 |
-6.95 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
-
37117478
-

-
37117479
-

-
45284557
-

-
34801270
-

-
34801271
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,3bR,9aR,9bR,11aS)-N,N-diisopropyl-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,6,8,9,9b,10,11-dodecah
(1S,3aR,3bR,9aR,9bR,11aS)-N,N-di…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
10.51 |
-11.56 |
1 |
4 |
0 |
49 |
400.607 |
3 |
↓
|
|
|
|
Analogs
-
36153213
-
-
36153217
-

-
36153221
-
-
35812109
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
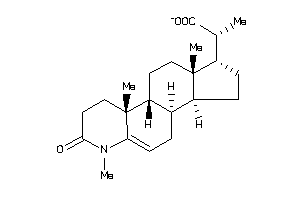
(2S)-2-[(1S,3aS,3bR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,8,9,9b,10,11-decahydro-1H-ind
(2S)-2-[(1S,3aS,3bR,9aR,9bR,11aS…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
11.1 |
-52.41 |
0 |
4 |
-1 |
60 |
358.502 |
2 |
↓
|
|
|
|
Analogs
-
36153217
-

-
36153221
-

-
36153209
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
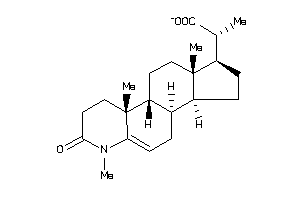
(2R)-2-[(1S,3aS,3bR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,8,9,9b,10,11-decahydro-1H-ind
(2R)-2-[(1S,3aS,3bR,9aR,9bR,11aS…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
10.8 |
-50.69 |
0 |
4 |
-1 |
60 |
358.502 |
2 |
↓
|
|
|
|
Analogs
-
36153221
-

-
36153209
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-2-[(1R,3aS,3bR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,8,9,9b,10,11-decahydro-1H-ind
(2S)-2-[(1R,3aS,3bR,9aR,9bR,11aS…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
11.07 |
-51.22 |
0 |
4 |
-1 |
60 |
358.502 |
2 |
↓
|
|
|
|
Analogs
-
36153209
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-2-[(1R,3aS,3bR,9aR,9bR,11aS)-6,9a,11a-trimethyl-7-oxo-2,3,3a,3b,4,8,9,9b,10,11-decahydro-1H-ind
(2R)-2-[(1R,3aS,3bR,9aR,9bR,11aS…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
11.07 |
-48.47 |
0 |
4 |
-1 |
60 |
358.502 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
0.94 |
-10.13 |
1 |
5 |
0 |
64 |
323.776 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
0.95 |
-10.2 |
1 |
5 |
0 |
64 |
323.776 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4R,9aS,9bS,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4R,9aS,9bS,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.63 |
-6.25 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4R,9aR,9bS,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4R,9aR,9bS,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.6 |
-6.1 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4R,9aS,9bR,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4R,9aS,9bR,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
9.99 |
-6.99 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4R,9aR,9bR,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4R,9aR,9bR,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
9.97 |
-6.93 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4S,9aS,9bS,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4S,9aS,9bS,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.5 |
-6.13 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4S,9aR,9bS,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4S,9aR,9bS,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.47 |
-6.02 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4S,9aS,9bR,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4S,9aS,9bR,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.02 |
-6.69 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,3bR,4S,9aR,9bR,11aR)-4,6,9a,11a-tetramethylspiro[2,3,3a,3b,4,8,9,9b,10,11-decahydroindeno[5,
(1R,3aS,3bR,4S,9aR,9bR,11aR)-4,6…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.05 |
-6.72 |
0 |
3 |
0 |
30 |
357.538 |
0 |
↓
|
|