|
|
Analogs
-
402782
-
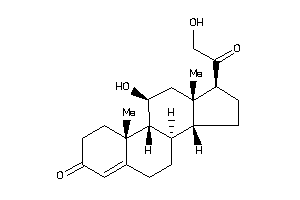
-
4095869
-
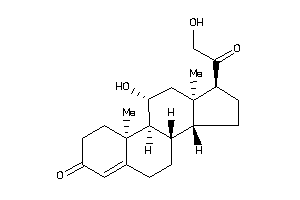
-
4095870
-
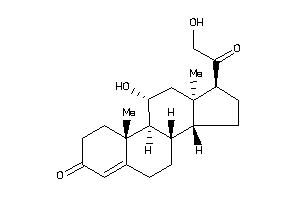
-
4095871
-
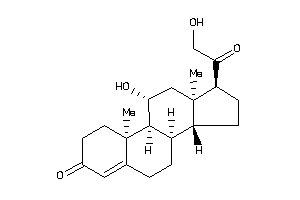
-
4095872
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
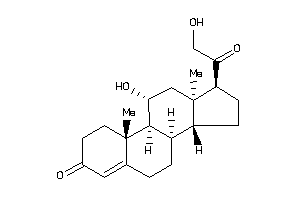
(8R,9R,10S,11R,13S,14S,17R)-17-glycoloyl-11-hydroxy-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15,16,17-dod
(8R,9R,10S,11R,13S,14S,17R)-17-g…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
-1.71 |
-10.96 |
2 |
4 |
0 |
74 |
346.467 |
2 |
↓
|
|
|
|
Analogs
-
119967
-
-
155356
-
-
525571
-
-
1069133
-
-
3881689
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
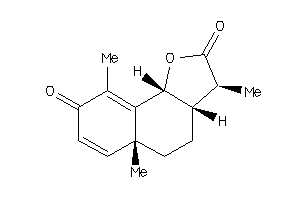
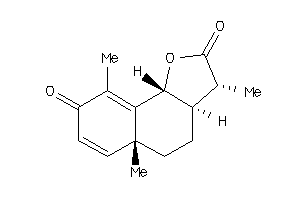
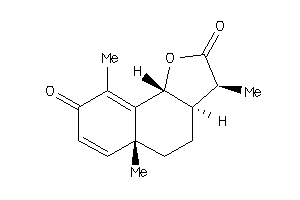
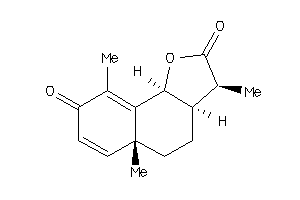
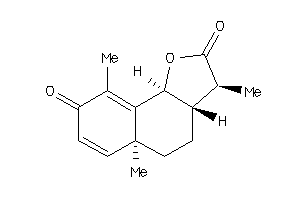
3,5a,9-trimethyl-3a,5,5a,9b-tetrahydronaphtho[1,2-b]furan-2,8(3H,4H)-dione
3,5a,9-trimethyl-3a,5,5a,9b-tetr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.84 |
-13.76 |
0 |
3 |
0 |
43 |
246.306 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
9.78 |
-7.88 |
0 |
5 |
0 |
54 |
344.407 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
10.05 |
-7.47 |
0 |
5 |
0 |
54 |
344.407 |
5 |
↓
|
|
|
|
Analogs
-
36371459
-
-
36371460
-
-
36371461
-
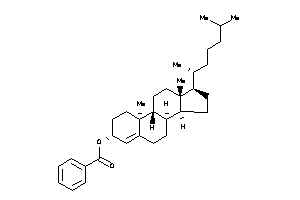
Draw
Identity
99%
90%
80%
70%
Popular Name:
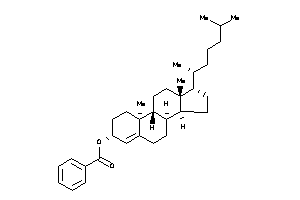
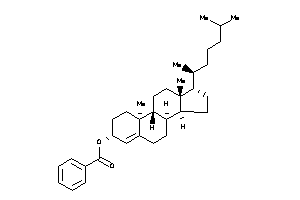
[(E,1R)-5-[(1R,2R,4aS,8aS)-2-hydroxy-2,5,5,8a-tetramethyl-decalin-1-yl]-1-[[(1S)-2,2-dimethyl-6-meth
[(E,1R)-5-[(1R,2R,4aS,8aS)-2-hyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.35 |
18.46 |
-6.93 |
1 |
3 |
0 |
47 |
548.852 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
9.62 |
-7.16 |
0 |
5 |
0 |
54 |
334.412 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
10.11 |
-6.03 |
0 |
5 |
0 |
54 |
334.412 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z)-[(1R,4aR,4bS,8aS,10aS)-1-[2-(3-furyl)ethyl]-4b,8,8,10a-tetramethyl-1,3,4,4a,5,6,7,8a,9,10-decah
[(Z)-[(1R,4aR,4bS,8aS,10aS)-1-[2…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
2.99 |
-7.36 |
4 |
8 |
0 |
121 |
537.767 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.10 |
3.64 |
-39.6 |
5 |
8 |
1 |
123 |
538.775 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4S,5aS,5bR,7aS,11aS,11bR,13aS,13bS)-1-acetoxy-5b,8,8,11a,13a-pentamethyl-13-oxo-4,5,5a,6,7,7a,9
[(1S,4S,5aS,5bR,7aS,11aS,11bR,13…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50378-1-O |
Mycobacterium Intracellulare (cluster #1 Of 1), Other |
Other |
5100 |
0.21 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50378 |
Z50378
|
Mycobacterium Intracellulare |
5100 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.61 |
13.66 |
-15.69 |
0 |
6 |
0 |
79 |
486.649 |
4 |
↓
|
|
|
|
Analogs
-
42852815
-
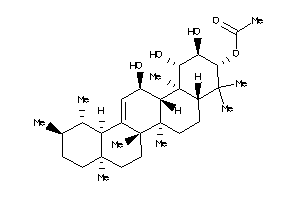
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,2R,3R,4aS,6aR,6bS,8aR,12aR,14R,14aS,14bS)-1,2,14-trihydroxy-4,4,6a,6b,8a,11,11,14b-octamethyl-1
[(1S,2R,3R,4aS,6aR,6bS,8aR,12aR,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.20 |
9.75 |
-8.31 |
3 |
5 |
0 |
87 |
516.763 |
2 |
↓
|
|
|
|
Analogs
-
5758835
-
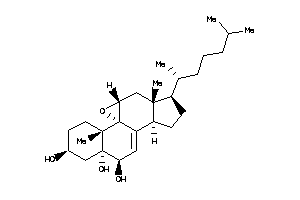
-
42877121
-
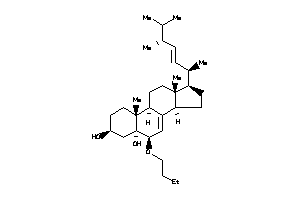
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,5R,6R,9S,10R,13R,14R,17R)-6-butoxy-17-[(E,1R,4R)-4-ethyl-1,5-dimethyl-hex-2-enyl]-10,13-dimethyl
(3S,5R,6R,9S,10R,13R,14R,17R)-6-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.06 |
12.67 |
-2.63 |
2 |
3 |
0 |
50 |
500.808 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
5.31 |
-22.07 |
2 |
9 |
0 |
136 |
502.56 |
4 |
↓
|
|
|
|
Analogs
-
5763636
-
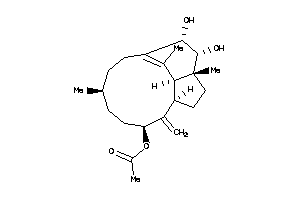
-
5763659
-
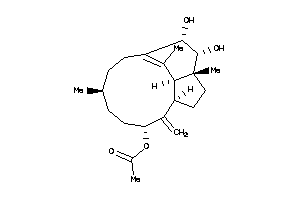
-
3652198
-
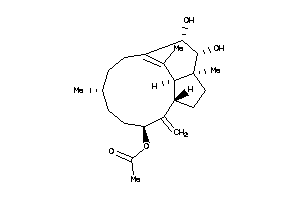
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2R,3R,6aR,6bS,8aR,11R,12S,12aR,14R,14aS,14bR)-3,14-dihydroxy-4,4,6a,6b,8a,11,12,14b-octamethyl-2,3
[(2R,3R,6aR,6bS,8aR,11R,12S,12aR…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.78 |
11.11 |
-6.98 |
2 |
4 |
0 |
67 |
498.748 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.55 |
16.33 |
-24.14 |
1 |
5 |
0 |
72 |
573.818 |
7 |
↓
|
|
|
|
Analogs
-
13704680
-
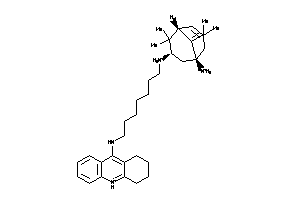
Draw
Identity
99%
90%
80%
70%
Popular Name:
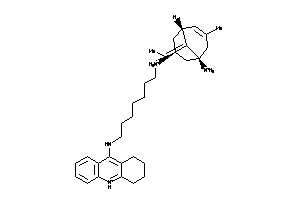
(1R,5R,7S,9E)-9-ethylidene-3,8,8-trimethyl-N7-[7-(1,2,3,4-tetrahydroacridin-9-ylamino)heptyl]bicyclo
(1R,5R,7S,9E)-9-ethylidene-3,8,8…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.35 |
15.41 |
-155.71 |
7 |
4 |
3 |
70 |
517.826 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
8.35 |
14.39 |
-73.99 |
6 |
4 |
2 |
66 |
516.818 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
8.35 |
15.18 |
-71.52 |
6 |
4 |
2 |
69 |
516.818 |
10 |
↓
|
|
|
|
Analogs
-
13704683
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.53 |
15.4 |
-148.91 |
7 |
4 |
3 |
70 |
489.772 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.53 |
15.17 |
-67.36 |
6 |
4 |
2 |
69 |
488.764 |
10 |
↓
|
|
|
|
Analogs
-
13704683
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.53 |
14.43 |
-153.11 |
7 |
4 |
3 |
70 |
489.772 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.53 |
14.2 |
-72.44 |
6 |
4 |
2 |
69 |
488.764 |
10 |
↓
|
|
|
|
Analogs
-
36455274
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.39 |
-16.22 |
3 |
6 |
0 |
104 |
480.601 |
4 |
↓
|
|
|
|
Analogs
-
43366257
-
Draw
Identity
99%
90%
80%
70%
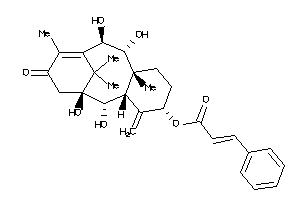
Popular Name:
3-[4-[[(7R,8R,9R,10R,13S,14S,17S)-17-acetyl-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodec
3-[4-[[(7R,8R,9R,10R,13S,14S,17S…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
12.4 |
-70.27 |
1 |
7 |
-1 |
116 |
633.856 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.07 |
12.09 |
-29.32 |
2 |
7 |
0 |
113 |
634.864 |
7 |
↓
|
|
|
|
Analogs
-
43366257
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[4-[[(7R,8S,9R,10R,13S,14S,17S)-17-acetyl-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodec
3-[4-[[(7R,8S,9R,10R,13S,14S,17S…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
12.53 |
-61.71 |
1 |
7 |
-1 |
116 |
633.856 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.07 |
12.28 |
-26.46 |
2 |
7 |
0 |
113 |
634.864 |
7 |
↓
|
|
|
|
Analogs
-
44201696
-
-
44201698
-
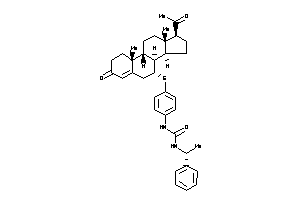
-
3965140
-
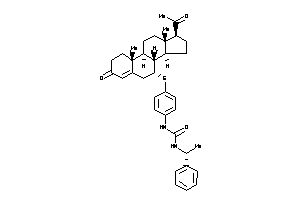
Draw
Identity
99%
90%
80%
70%
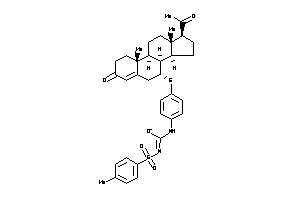
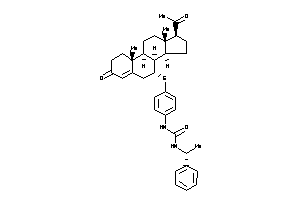
Popular Name:
3-[4-[[(7R,8S,9R,10R,13S,14S,17S)-17-acetyl-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodec
3-[4-[[(7R,8S,9R,10R,13S,14S,17S…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81008-1-O |
Human Breast Cancer Cell Lines (cluster #1 Of 1), Other |
Other |
800 |
0.20 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81008 |
Z81008
|
Human Breast Cancer Cell Lines |
600 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.05 |
16.58 |
-22.98 |
2 |
5 |
0 |
75 |
584.826 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14S,17S)-17-acetyl-13,17-dimethyl-1,2,6,7,8,9,10,11,12,14,15,16-dodecahydrocyclopenta
(8S,9R,10R,13S,14S,17S)-17-acety…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
10.57 |
-10.83 |
0 |
2 |
0 |
34 |
314.469 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aS)-5-[2-(5,8-diacetoxy-1,4-dihydronaphthalen-2-yl)ethyl]-1,4a-dimethyl-6-methylene-dec
[(1S,4aR,5S,8aS)-5-[2-(5,8-diace…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.05 |
19.05 |
-17.07 |
0 |
6 |
0 |
79 |
522.682 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aR)-5-[2-(5,8-diacetoxy-1,4-dihydronaphthalen-2-yl)ethyl]-1,4a-dimethyl-6-methylene-dec
[(1S,4aR,5S,8aR)-5-[2-(5,8-diace…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.05 |
18.61 |
-17.42 |
0 |
6 |
0 |
79 |
522.682 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
13.63 |
-9.27 |
0 |
4 |
0 |
60 |
420.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
13.66 |
-9.88 |
0 |
4 |
0 |
60 |
420.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aS)-5-[2-[(4aR,9aR)-9,10-dioxo-1,4,4a,9a-tetrahydroanthracen-2-yl]ethyl]-1,4a-dimethyl-
[(1S,4aR,5S,8aS)-5-[2-[(4aR,9aR)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.87 |
16.92 |
-10 |
0 |
4 |
0 |
60 |
488.668 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aR)-5-[2-[(4aR,9aR)-9,10-dioxo-1,4,4a,9a-tetrahydroanthracen-2-yl]ethyl]-1,4a-dimethyl-
[(1S,4aR,5S,8aR)-5-[2-[(4aR,9aR)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.87 |
16.49 |
-10.29 |
0 |
4 |
0 |
60 |
488.668 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
15.58 |
-18.71 |
0 |
7 |
0 |
91 |
524.654 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
7.81 |
-16.81 |
2 |
5 |
0 |
79 |
440.58 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
7.78 |
-16.86 |
2 |
5 |
0 |
79 |
440.58 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(7R,8S,9S,10R,13S,14S,17S)-17-hydroxy-7,13,17-trimethyl-1,2,6,7,8,9,10,11,12,14,15,16-dodecahydrocyc
(7R,8S,9S,10R,13S,14S,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
7.8 |
-8.16 |
1 |
2 |
0 |
37 |
302.458 |
0 |
↓
|
|
|
|
Analogs
-
6778072
-
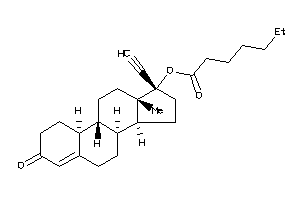
-
6778073
-
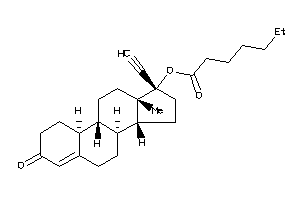
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.37 |
15.51 |
-11.96 |
0 |
3 |
0 |
43 |
410.598 |
7 |
↓
|
|
|
|
Analogs
-
31284427
-
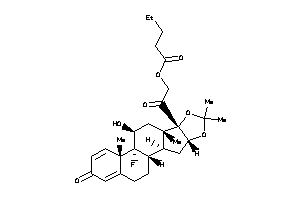
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
9.37 |
-14.1 |
1 |
7 |
0 |
99 |
476.541 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8S,9S,10S,13S,14S)-13-methyl-3,17-dioxo-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phenanthren
[(8S,9S,10S,13S,14S)-13-methyl-3…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
9.65 |
-15.23 |
0 |
4 |
0 |
60 |
328.408 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,3'R,3'aS,6'S,6aS,6bS,7'aR,9R,11aS,11bS)-3',6',10,11b-tetramethylspiro[2,3,4,6,6a,6b,7,8,11,11a-d
(3S,3'R,3'aS,6'S,6aS,6bS,7'aR,9R…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
9.69 |
-47.21 |
3 |
3 |
1 |
46 |
412.638 |
0 |
↓
|
|
|
|
Analogs
-
33830066
-
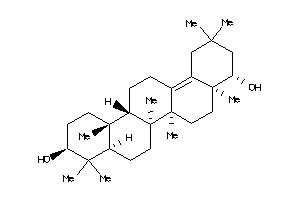
-
33830067
-
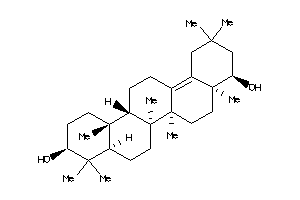
-
36375258
-
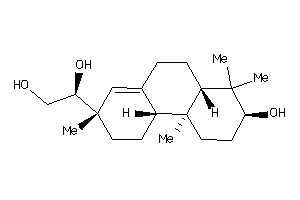
-
36375260
-
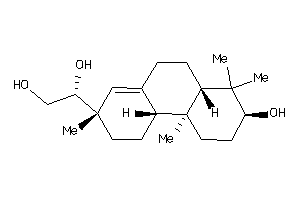
-
33754089
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4aR,6aR,6bS,8S,8aS,14aR,14bR)-4,4,6a,6b,8a,11,11,14b-octamethyl-1,2,3,4a,5,6,7,8,9,10,12,13,14,1
(3S,4aR,6aR,6bS,8S,8aS,14aR,14bR…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.95 |
9.31 |
-2.92 |
2 |
2 |
0 |
40 |
442.728 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
5.23 |
-7.9 |
1 |
2 |
0 |
37 |
232.323 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
6.69 |
-75.85 |
2 |
6 |
-1 |
115 |
393.5 |
8 |
↓
|
|
|
|
Analogs
-
28569326
-
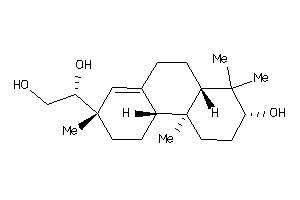
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,8S,9S,10R,14S)-10,17-dimethyl-1,2,3,6,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthren-3
(3R,8S,9S,10R,14S)-10,17-dimethy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
8.31 |
-2.42 |
1 |
1 |
0 |
20 |
272.432 |
0 |
↓
|
|
|
|
Analogs
-
28569326
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,8S,9S,10R,14S)-10,17-dimethyl-1,2,3,6,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthren-3
(3S,8S,9S,10R,14S)-10,17-dimethy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
8.25 |
-2.61 |
1 |
1 |
0 |
20 |
272.432 |
0 |
↓
|
|
|
|
Analogs
-
35415703
-
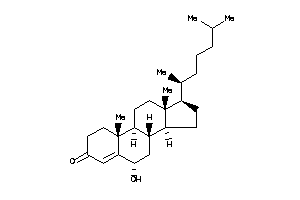
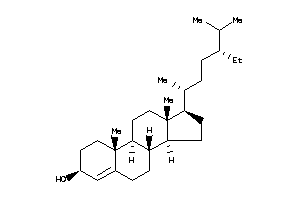
-
35415704
-
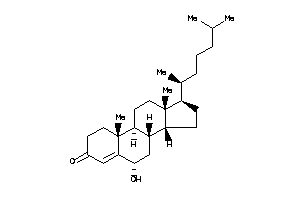
-
39012979
-
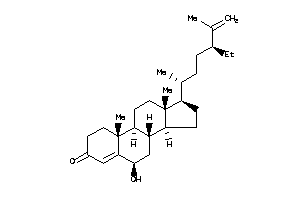
-
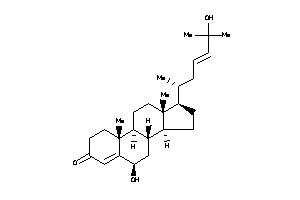
40395109
-
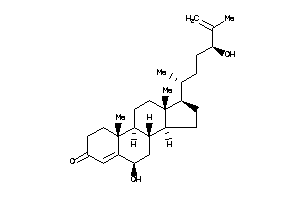
-
40848053
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6R,8S,9S,10R,14S)-6-hydroxy-10,17-dimethyl-2,6,7,8,9,11,12,14,15,16-decahydro-1H-cyclopenta[a]phena
(6R,8S,9S,10R,14S)-6-hydroxy-10,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
7.65 |
-6.48 |
1 |
2 |
0 |
37 |
286.415 |
0 |
↓
|
|
|
|
Analogs
-
5758815
-
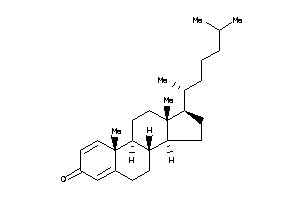
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10R,13S,14S)-10,13-dimethyl-17-methylene-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phena
(8S,9S,10R,13S,14S)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
10.57 |
-7.97 |
0 |
1 |
0 |
17 |
282.427 |
0 |
↓
|
|
|
|
Analogs
-
12496631
-
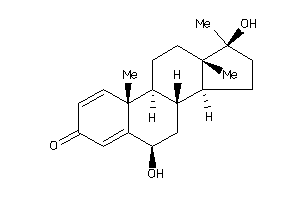
Draw
Identity
99%
90%
80%
70%
Popular Name:
(6R,8S,9S,10R,13S,14S)-6-hydroxy-10,13-dimethyl-17-methylene-7,8,9,11,12,14,15,16-octahydro-6H-cyclo
(6R,8S,9S,10R,13S,14S)-6-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
7.6 |
-6.98 |
1 |
2 |
0 |
37 |
298.426 |
0 |
↓
|
|
|
|
Analogs
-
12496631
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(6R,8S,9S,10R,13R,14S)-6-hydroxy-10,13-dimethyl-6,7,8,9,11,12,14,15-octahydrocyclopenta[a]phenanthre
(6R,8S,9S,10R,13R,14S)-6-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
7.04 |
-7.04 |
1 |
2 |
0 |
37 |
284.399 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9S,10R,13R,14S)-13-methyl-2,6,7,8,9,10,11,12,14,15-decahydro-1H-cyclopenta[a]phenanthren-3-one
(8R,9S,10R,13R,14S)-13-methyl-2,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
10.05 |
-6.42 |
0 |
1 |
0 |
17 |
256.389 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,13R,14S)-13-methyl-2,6,7,8,11,12,14,15-octahydro-1H-cyclopenta[a]phenanthren-3-one
(8S,13R,14S)-13-methyl-2,6,7,8,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
10.07 |
-6.7 |
0 |
1 |
0 |
17 |
254.373 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9S,10R,13S,14S)-13-methyl-17-methylene-1,2,6,7,8,9,10,11,12,14,15,16-dodecahydrocyclopenta[a]phe
(8R,9S,10R,13S,14S)-13-methyl-17…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.23 |
10.56 |
-6.46 |
0 |
1 |
0 |
17 |
270.416 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(9S,13S,14R)-13-methyl-6,9,11,12,14,15,16,17-octahydrocyclopenta[a]phenanthren-3-ol
(9S,13S,14R)-13-methyl-6,9,11,12…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
8.04 |
-3.44 |
1 |
1 |
0 |
20 |
254.373 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10R,13S,14S,17S)-N-[1-(4-chlorophenyl)cyclopentyl]-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,
(8S,9S,10R,13S,14S,17S)-N-[1-(4-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.11 |
15.03 |
-11.75 |
1 |
3 |
0 |
46 |
494.119 |
3 |
↓
|
|