|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
36160273
-
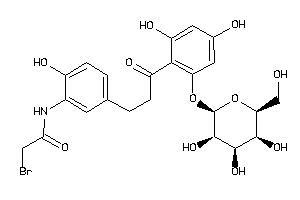
-
36160277
-
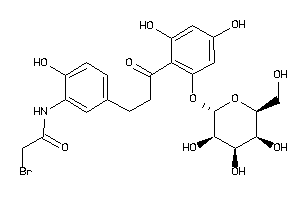
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.20 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1_HUMAN |
P13866
|
Sodium/glucose Cotransporter 1, Human |
7700 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.51 |
-6.67 |
-23.03 |
8 |
12 |
0 |
206 |
572.361 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.51 |
-5.94 |
-56.96 |
7 |
12 |
-1 |
209 |
571.353 |
9 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
36160291
-
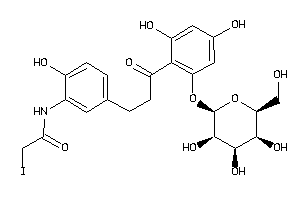
-
36160295
-
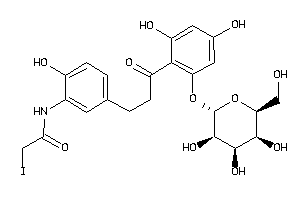
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.20 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1_HUMAN |
P13866
|
Sodium/glucose Cotransporter 1, Human |
7000 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.78 |
-6.19 |
-22.16 |
8 |
12 |
0 |
206 |
619.361 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.78 |
-5.47 |
-55.84 |
7 |
12 |
-1 |
209 |
618.353 |
9 |
↓
|
|
|
|
Analogs
-
36160312
-
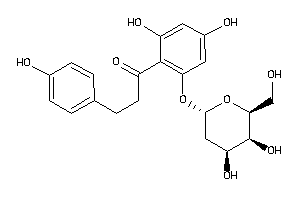
-
29407645
-
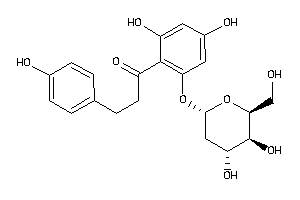
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.26 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1_HUMAN |
P13866
|
Sodium/glucose Cotransporter 1, Human |
2600 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
-4 |
-18.71 |
6 |
9 |
0 |
157 |
420.414 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.10 |
-3.27 |
-50.89 |
5 |
9 |
-1 |
160 |
419.406 |
7 |
↓
|
|
|
|
Analogs
-
36160308
-
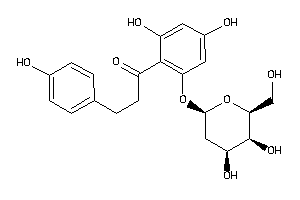
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.26 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1_HUMAN |
P13866
|
Sodium/glucose Cotransporter 1, Human |
2600 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
-3.95 |
-16.13 |
6 |
9 |
0 |
157 |
420.414 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.10 |
-3.22 |
-47.44 |
5 |
9 |
-1 |
160 |
419.406 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
36160316
-
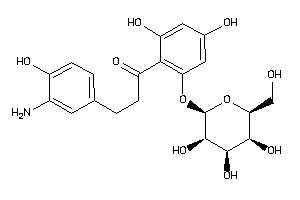
-
29409150
-
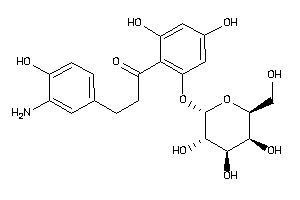
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1_HUMAN |
P13866
|
Sodium/glucose Cotransporter 1, Human |
3800 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.07 |
-7.69 |
-17.47 |
9 |
11 |
0 |
203 |
451.428 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.07 |
-6.97 |
-48.38 |
8 |
11 |
-1 |
206 |
450.42 |
7 |
↓
|
|
|
|
Analogs
-
3874668
-
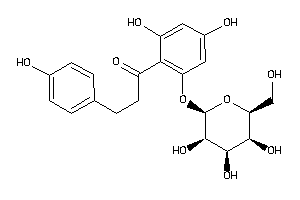
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[2,4-dihydroxy-6-[(2R,3R,4R,5S,6S)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydropyran-2-yl]oxy-phen
1-[2,4-dihydroxy-6-[(2R,3R,4R,5S…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1_HUMAN |
P13866
|
Sodium/glucose Cotransporter 1, Human |
2800 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
-3.4 |
-17.73 |
6 |
10 |
0 |
166 |
450.44 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.94 |
-2.67 |
-48.72 |
5 |
10 |
-1 |
169 |
449.432 |
8 |
↓
|
|
|
|
Analogs
-
3874668
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[2,4-dihydroxy-6-[(2S,3R,4R,5S,6S)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydropyran-2-yl]oxy-phen
1-[2,4-dihydroxy-6-[(2S,3R,4R,5S…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1-1-E |
Sodium/glucose Cotransporter 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC5A1_HUMAN |
P13866
|
Sodium/glucose Cotransporter 1, Human |
2800 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
-3.53 |
-17.01 |
6 |
10 |
0 |
166 |
450.44 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.94 |
-2.8 |
-47.01 |
5 |
10 |
-1 |
169 |
449.432 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
-4.04 |
-15.44 |
6 |
9 |
0 |
157 |
420.414 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.40 |
-6.89 |
-17.03 |
7 |
10 |
0 |
177 |
436.413 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[2-[(2R,3R,4R,5R,6S)-3,5-dihydroxy-6-(hydroxymethyl)-4-methoxy-tetrahydropyran-2-yl]oxy-4,6-dihydr
1-[2-[(2R,3R,4R,5R,6S)-3,5-dihyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
-3.3 |
-21.69 |
6 |
10 |
0 |
166 |
450.44 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.80 |
-2.57 |
-50.9 |
5 |
10 |
-1 |
169 |
449.432 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[2-[(2R,3R,4S,5R,6S)-3,5-dihydroxy-6-(hydroxymethyl)-4-methoxy-tetrahydropyran-2-yl]oxy-4,6-dihydr
1-[2-[(2R,3R,4S,5R,6S)-3,5-dihyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
-3.84 |
-19.16 |
6 |
10 |
0 |
166 |
450.44 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.80 |
-3.11 |
-49.3 |
5 |
10 |
-1 |
169 |
449.432 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[2-[(2R,3R,4R,5S,6S)-3,5-dihydroxy-6-(hydroxymethyl)-4-methoxy-tetrahydropyran-2-yl]oxy-4,6-dihydr
1-[2-[(2R,3R,4R,5S,6S)-3,5-dihyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
-4.4 |
-19.64 |
6 |
10 |
0 |
166 |
450.44 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.80 |
-3.68 |
-47.54 |
5 |
10 |
-1 |
169 |
449.432 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[2-[(2R,3R,4S,5S,6S)-3,5-dihydroxy-6-(hydroxymethyl)-4-methoxy-tetrahydropyran-2-yl]oxy-4,6-dihydr
1-[2-[(2R,3R,4S,5S,6S)-3,5-dihyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
-4.53 |
-18.28 |
6 |
10 |
0 |
166 |
450.44 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.80 |
-3.8 |
-47.25 |
5 |
10 |
-1 |
169 |
449.432 |
8 |
↓
|
|