|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
9.24 |
-12.92 |
0 |
4 |
0 |
41 |
387.277 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
469387
-
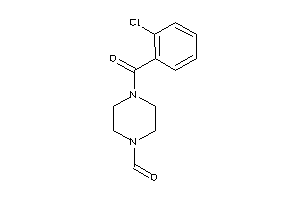
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
-0.22 |
-14.38 |
0 |
4 |
0 |
41 |
328.799 |
2 |
↓
|
|
|
|
Analogs
-
19773592
-
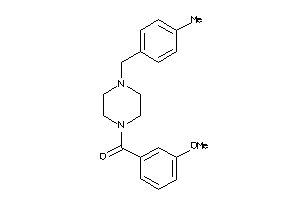
-
30896018
-
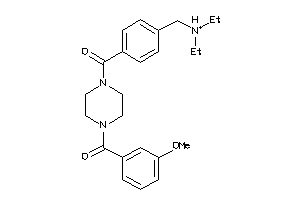
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
7.95 |
-16.14 |
0 |
5 |
0 |
50 |
338.407 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
18840911
-
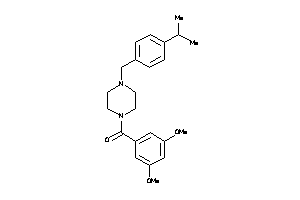
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
8.48 |
-14.65 |
0 |
6 |
0 |
59 |
396.487 |
5 |
↓
|
|
|
|
Analogs
-
25516940
-
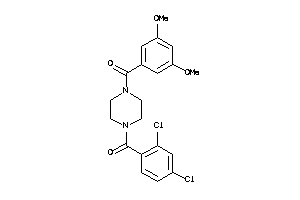
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
7.83 |
-17.14 |
0 |
5 |
0 |
50 |
358.825 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
7.64 |
-21.26 |
0 |
6 |
0 |
74 |
349.39 |
3 |
↓
|
|
|
|
Analogs
-
23791000
-
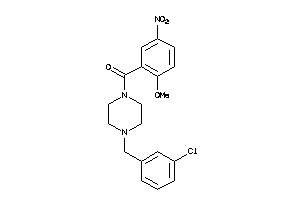
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
8.94 |
-15.99 |
0 |
8 |
0 |
96 |
403.822 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
7.95 |
-16.01 |
0 |
7 |
0 |
78 |
463.987 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
9.6 |
-11.62 |
0 |
4 |
0 |
41 |
358.388 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
7.64 |
-17.75 |
1 |
5 |
0 |
61 |
352.434 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.19 |
8.48 |
-55.54 |
0 |
5 |
-1 |
64 |
351.426 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
6.93 |
-21.84 |
0 |
6 |
0 |
75 |
400.5 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
10.12 |
-14.04 |
0 |
4 |
0 |
41 |
401.304 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
13010223
-
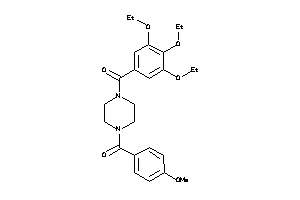
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
7.03 |
-17.62 |
1 |
8 |
0 |
89 |
442.512 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-Piperazinecarboxylic acid, 1,4-bis(3,4,5-trimethoxybenzoyl)-, ethyl ester; Ethyl 1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinecarboxylate; LS-110830; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-ethoxycarbonylpiperazine
2-Piperazinecarboxylic acid, 1,4…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
8.88 |
-22.35 |
0 |
12 |
0 |
122 |
546.573 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-Piperazinecarboxylic acid, 1,4-bis(3,4,5-trimethoxybenzoyl)-, hexyl ester; Hexyl 1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinecarboxylate; LS-110831; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-n-hexyloxycarbonyl piperazine
2-Piperazinecarboxylic acid, 1,4…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
12.13 |
-18.71 |
0 |
12 |
0 |
122 |
602.681 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1,4-Bis(3,4,5-trimethoxybenzoyl)-N-hexyl-2-piperazinecarboxamide; 2-Piperazinecarboxamide, 1,4-bis(3,4,5-trimethoxybenzoyl)-N-hexyl-; LS-110654; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-(N-m-hexyl)amidopiperazine
1,4-Bis(3,4,5-trimethoxybenzoyl)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
9.54 |
-20.69 |
1 |
12 |
0 |
125 |
601.697 |
14 |
↓
|
|
|
|
Analogs
-
3914859
-
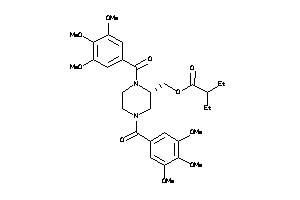
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl 2-ethylbutanoate; Butanoic acid, 2-ethyl-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-46283; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-(2'-ethyl)butanoyloxymethylpiperazine
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.23 |
Binding ≤ 10μM
|
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
11.05 |
-22.86 |
0 |
12 |
0 |
122 |
602.681 |
14 |
↓
|
|
|
|
Analogs
-
3914861
-
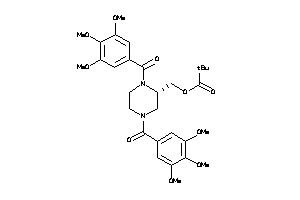
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl 2,2-dimethylpropanoate; LS-121402; N,N'-Bis(3,4,5-trimethoxylbenzoyl)-2-piperizinylmethyl 2,2-dimethylpropanoate; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-tert-butylcarbonyloxymethylpiperazine; Propanoi
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.21 |
Binding ≤ 10μM
|
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.23 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
100 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
10.28 |
-20.68 |
0 |
12 |
0 |
122 |
588.654 |
12 |
↓
|
|
|
|
Analogs
-
14243288
-
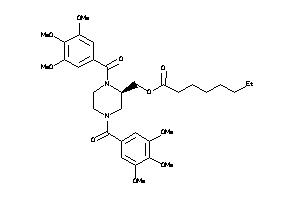
-
25962869
-
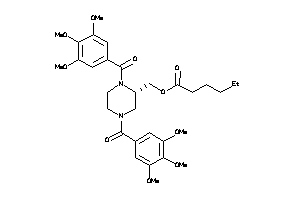
-
25962873
-
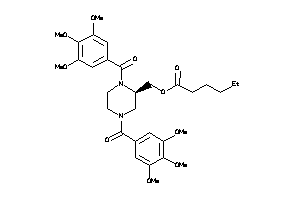
-
3935342
-
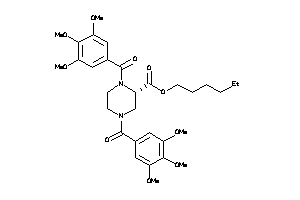
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl octanoate; LS-97952; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-n-octanoyloxymethylpiperazine; Octanoic acid, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
310 |
0.20 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
310 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
13.26 |
-20.68 |
0 |
12 |
0 |
122 |
630.735 |
17 |
↓
|
|
|
|
Analogs
-
25962869
-

-
25962873
-

-
3935342
-

-
14243287
-
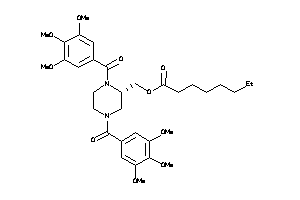
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl octanoate; LS-97952; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-n-octanoyloxymethylpiperazine; Octanoic acid, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
310 |
0.20 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
310 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
12.95 |
-20.61 |
0 |
12 |
0 |
122 |
630.735 |
17 |
↓
|
|
|
|
Analogs
-
3914863
-
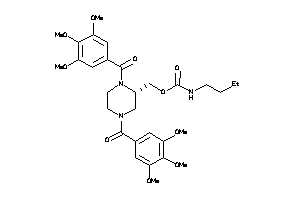
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl butylcarbamate; Carbamic acid, butyl-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-49075; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N"-(n-butyl)-carbamoyloxymethylpiperazine
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
7.56 |
-21.4 |
1 |
13 |
0 |
134 |
603.669 |
14 |
↓
|
|
|
|
Analogs
-
3914864
-
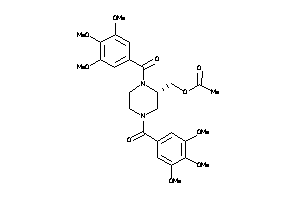
Draw
Identity
99%
90%
80%
70%
Popular Name:
1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinemethanol acetate (ester); 2-Piperazinemethanol, 1,4-bis(3,4,5-trimethoxybenzoyl)-, acetate (ester); LS-112740; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-acetyloxymethylpiperazine
1,4-Bis(3,4,5-trimethoxybenzoyl)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.23 |
Binding ≤ 10μM
|
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
8.3 |
-21.31 |
0 |
12 |
0 |
122 |
546.573 |
11 |
↓
|
|
|
|
Analogs
-
3914865
-
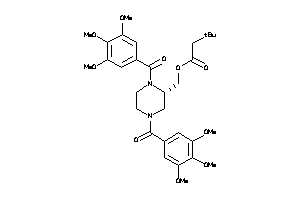
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl 3,3-dimethylbutanoate; Butanoic acid, 3,3-dimethyl-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-46261; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-tert-butyl acetyloxymethylpiperazine
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.23 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
120 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
10.76 |
-19.62 |
0 |
12 |
0 |
122 |
602.681 |
13 |
↓
|
|
|
|
Analogs
-
14243294
-
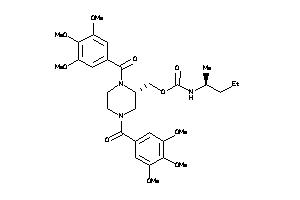
-
14243295
-
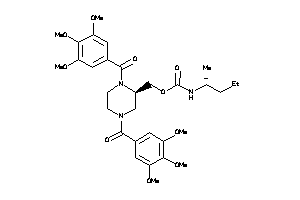
-
14243296
-
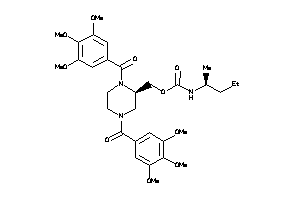
-
3914868
-
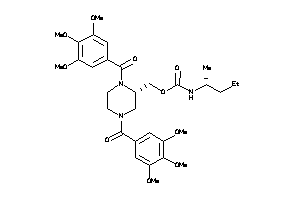
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (1-ethylpropyl)carbamate; Carbamic acid, (1-ethylpropyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-49771; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(1'-ethyl)propylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
130 |
0.22 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
130 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
8.44 |
-20.76 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
Analogs
-
3914867
-
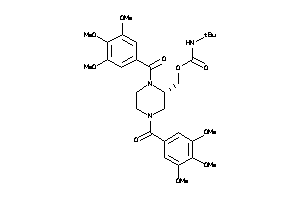
Draw
Identity
99%
90%
80%
70%
Popular Name:
Carbamic acid, (1,1-dimethylethyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-49556; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-tert-butylcarbamoyloxymethylpiperazine
Carbamic acid, (1,1-dimethylethy…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
130 |
0.22 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
130 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
7.46 |
-24.33 |
1 |
13 |
0 |
134 |
603.669 |
12 |
↓
|
|
|
|
Analogs
-
14243295
-

-
14243296
-

-
3914868
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (1-methylbutyl)carbamate; Carbamic acid, (1-methylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-50071; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(1'-methyl)butylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.22 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
100 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
8.97 |
-21.21 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
Analogs
-
14243296
-

-
3914868
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (1-methylbutyl)carbamate; Carbamic acid, (1-methylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-50071; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(1'-methyl)butylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.22 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
100 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
8.67 |
-21.38 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
Analogs
-
3914868
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (1-methylbutyl)carbamate; Carbamic acid, (1-methylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-50071; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(1'-methyl)butylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.22 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
100 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
8.67 |
-21.28 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3914869
-
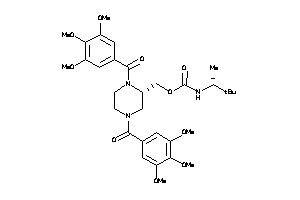
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
170 |
0.21 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
170 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
9.19 |
-24.46 |
1 |
13 |
0 |
134 |
631.723 |
13 |
↓
|
|
|
|
Analogs
-
3914870
-
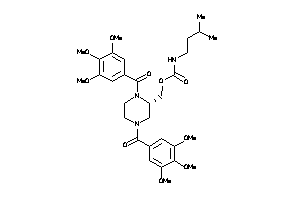
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (3-methylbutyl)carbamate; Carbamic acid, (3-methylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-50073; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(3'-methyl)butylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
140 |
0.22 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
140 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
8.31 |
-24.59 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
Analogs
-
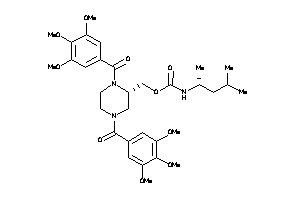
14243302
-
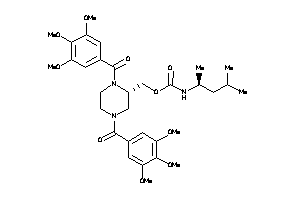
-
14243303
-
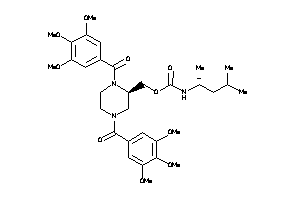
-
3914871
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
Carbamic acid, (1,3-dimethylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-49450; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(1',3'-dimethyl)butylcarbamoyloxymethylpiperazine
Carbamic acid, (1,3-dimethylbuty…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
180 |
0.21 |
Binding ≤ 10μM
|
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
280 |
0.20 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
280 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
9.37 |
-21.33 |
1 |
13 |
0 |
134 |
631.723 |
14 |
↓
|
|
|
|
Analogs
-
14243303
-

-
3914871
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
Carbamic acid, (1,3-dimethylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-49450; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(1',3'-dimethyl)butylcarbamoyloxymethylpiperazine
Carbamic acid, (1,3-dimethylbuty…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
180 |
0.21 |
Binding ≤ 10μM
|
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
280 |
0.20 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
280 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
9.5 |
-22.12 |
1 |
13 |
0 |
134 |
631.723 |
14 |
↓
|
|
|
|
Analogs
-
3914871
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
Carbamic acid, (1,3-dimethylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-49450; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(1',3'-dimethyl)butylcarbamoyloxymethylpiperazine
Carbamic acid, (1,3-dimethylbuty…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
180 |
0.21 |
Binding ≤ 10μM
|
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
280 |
0.20 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
280 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
9.44 |
-21.4 |
1 |
13 |
0 |
134 |
631.723 |
14 |
↓
|
|
|
|
Analogs
-
14243305
-
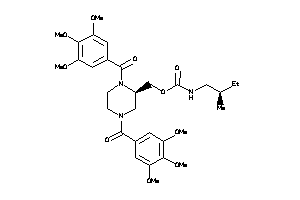
-
14243306
-
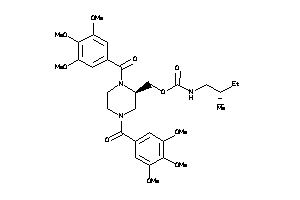
-
3914872
-
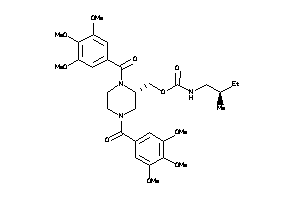
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (2-methylbutyl)carbamate; Carbamic acid, (2-methylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-50072; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(2'-methyl)butylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
470 |
0.20 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
470 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
8.52 |
-21.04 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
Analogs
-
14243306
-

-
3914872
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (2-methylbutyl)carbamate; Carbamic acid, (2-methylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-50072; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(2'-methyl)butylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
470 |
0.20 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
470 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
8.22 |
-21.19 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
Analogs
-
3914872
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1,4-Bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl (2-methylbutyl)carbamate; Carbamic acid, (2-methylbutyl)-, (1,4-bis(3,4,5-trimethoxybenzoyl)-2-piperazinyl)methyl ester; LS-50072; N,N'-Di-(3',4',5'-trimethoxybenzoyl)-2-N''-(2'-methyl)butylcarbamoylo
(1,4-Bis(3,4,5-trimethoxybenzoyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
470 |
0.20 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR_HUMAN |
P25105
|
Platelet Activating Factor Receptor, Human |
470 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
8.47 |
-24.39 |
1 |
13 |
0 |
134 |
617.696 |
14 |
↓
|
|
|
|
Analogs
-
53152251
-
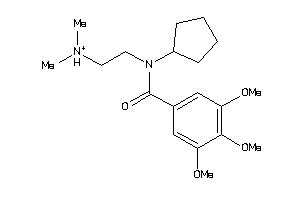
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
12.28 |
-18.13 |
0 |
10 |
0 |
96 |
586.726 |
15 |
↓
|
|
|
|
Analogs
-
53152251
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
11.96 |
-18.66 |
0 |
10 |
0 |
96 |
586.726 |
15 |
↓
|
|
|
|
Analogs
-
53152251
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
9.74 |
-18.89 |
0 |
10 |
0 |
96 |
544.645 |
12 |
↓
|
|