|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
230 |
0.23 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
12.29 |
-29.31 |
1 |
13 |
0 |
161 |
578.511 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
230 |
0.23 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
700 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
12.28 |
-32.52 |
1 |
13 |
0 |
161 |
578.511 |
12 |
↓
|
|
|
|
Analogs
-
6522938
-
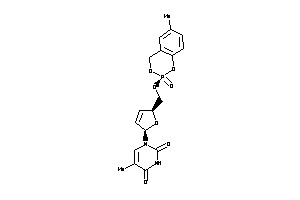
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4600 |
0.27 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
460 |
0.32 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
460 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
6.3 |
-17.24 |
1 |
9 |
0 |
109 |
406.331 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4600 |
0.27 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
460 |
0.32 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
460 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
6.3 |
-18.58 |
1 |
9 |
0 |
109 |
406.331 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
7.05 |
-17.38 |
2 |
9 |
0 |
114 |
361.298 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.05 |
7.3 |
-40.16 |
3 |
9 |
1 |
116 |
362.306 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.05 |
2.9 |
-37.61 |
3 |
9 |
1 |
116 |
362.306 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
7.12 |
-17.11 |
2 |
9 |
0 |
114 |
361.298 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.05 |
7.37 |
-37.15 |
3 |
9 |
1 |
116 |
362.306 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.05 |
2.97 |
-34.3 |
3 |
9 |
1 |
116 |
362.306 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
7.76 |
-18.04 |
2 |
9 |
0 |
114 |
375.325 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.45 |
8.01 |
-41.55 |
3 |
9 |
1 |
116 |
376.333 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.45 |
3.6 |
-39.22 |
3 |
9 |
1 |
116 |
376.333 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
7.76 |
-17.59 |
2 |
9 |
0 |
114 |
375.325 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.45 |
8.01 |
-37.73 |
3 |
9 |
1 |
116 |
376.333 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.45 |
3.61 |
-35.02 |
3 |
9 |
1 |
116 |
376.333 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
3000 |
0.27 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
4500 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
9.34 |
-17.48 |
2 |
9 |
0 |
114 |
417.406 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.71 |
9.59 |
-41.07 |
3 |
9 |
1 |
116 |
418.414 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.71 |
5.17 |
-38.71 |
3 |
9 |
1 |
116 |
418.414 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
3000 |
0.27 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
4500 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
9.33 |
-16.89 |
2 |
9 |
0 |
114 |
417.406 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.71 |
9.58 |
-36.99 |
3 |
9 |
1 |
116 |
418.414 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.71 |
5.17 |
-34.2 |
3 |
9 |
1 |
116 |
418.414 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
5500 |
0.22 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
5300 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
11.68 |
-16.45 |
2 |
9 |
0 |
114 |
473.514 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.37 |
11.93 |
-40.32 |
3 |
9 |
1 |
116 |
474.522 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.37 |
7.52 |
-38.03 |
3 |
9 |
1 |
116 |
474.522 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
5500 |
0.22 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
5300 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
11.66 |
-16.07 |
2 |
9 |
0 |
114 |
473.514 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.37 |
11.91 |
-36.48 |
3 |
9 |
1 |
116 |
474.522 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.37 |
7.51 |
-33.65 |
3 |
9 |
1 |
116 |
474.522 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80737-1-O |
C3H/3T3 (Embryonic Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
1700 |
0.28 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
4530 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
8.52 |
-20.28 |
2 |
10 |
0 |
124 |
421.325 |
4 |
↓
|
|
|
|
Analogs
-
40786174
-
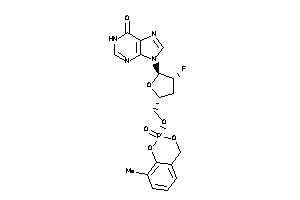
-
40786176
-
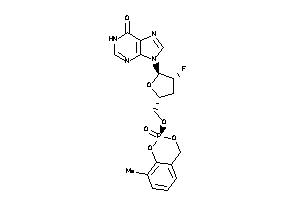
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
3570 |
0.25 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874 |
Z80874
|
CEM (T-cell Leukemia) |
3570 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
9.08 |
-32.56 |
1 |
10 |
0 |
118 |
436.336 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.29 |
7.01 |
-66.51 |
0 |
10 |
-1 |
121 |
435.328 |
4 |
↓
|
|
|
|
Analogs
-
40786174
-

-
40786176
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
3570 |
0.25 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874 |
Z80874
|
CEM (T-cell Leukemia) |
3570 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
9.08 |
-33 |
1 |
10 |
0 |
118 |
436.336 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.29 |
7.01 |
-67.97 |
0 |
10 |
-1 |
121 |
435.328 |
4 |
↓
|
|