|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-1-[[(1R,4aR,8aS)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methyl]-4-hydrox
[(E,1S)-1-[[(1R,4aR,8aS)-2,5,5,8…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.70 |
9.5 |
-8.16 |
1 |
3 |
0 |
47 |
348.527 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-1-[[(1R,4aR,8aR)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methyl]-4-hydrox
[(E,1S)-1-[[(1R,4aR,8aR)-2,5,5,8…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.70 |
9.22 |
-7.91 |
1 |
3 |
0 |
47 |
348.527 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-1-[[(1S,4aR,8aS)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methyl]-4-hydrox
[(E,1S)-1-[[(1S,4aR,8aS)-2,5,5,8…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.70 |
9.75 |
-8.05 |
1 |
3 |
0 |
47 |
348.527 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-1-[[(1S,4aR,8aR)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methyl]-4-hydrox
[(E,1S)-1-[[(1S,4aR,8aR)-2,5,5,8…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.70 |
9.73 |
-7.83 |
1 |
3 |
0 |
47 |
348.527 |
6 |
↓
|
|
|
|
Analogs
-
704422
-
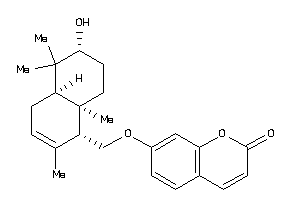
-
704423
-
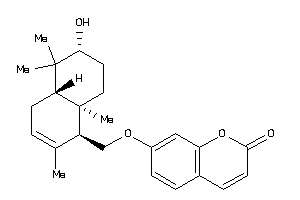
-
704424
-
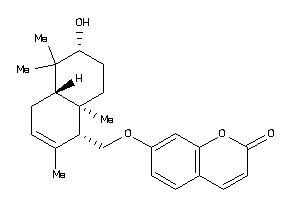
Draw
Identity
99%
90%
80%
70%
Popular Name:
7-[[(1S,4aR,6R,8aS)-6-hydroxy-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methoxy]chr
7-[[(1S,4aR,6R,8aS)-6-hydroxy-2,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
10.18 |
-14.53 |
1 |
4 |
0 |
60 |
382.5 |
3 |
↓
|
|
|
|
Analogs
-
4026022
-
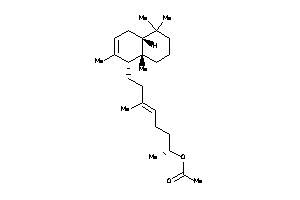
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-7-[(1R,4aR,8aR)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]-1,5-dimethyl-hep
[(E,1S)-7-[(1R,4aR,8aR)-2,5,5,8a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.75 |
15.3 |
-5 |
0 |
2 |
0 |
26 |
374.609 |
8 |
↓
|
|
|
|
Analogs
-
4026022
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-7-[(1S,4aR,8aR)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]-1,5-dimethyl-hep
[(E,1S)-7-[(1S,4aR,8aR)-2,5,5,8a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.75 |
15.35 |
-4.82 |
0 |
2 |
0 |
26 |
374.609 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R)-1-[[(1S,4aS,8aS)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methyl]-5-(3-furyl
[(1R)-1-[[(1S,4aS,8aS)-2,5,5,8a-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-2-E |
Dual Specificity Protein Phosphatase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4600 |
0.25 |
Binding ≤ 10μM |
|
MPIP1-1-E |
Dual Specificity Phosphatase Cdc25A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.25 |
Binding ≤ 10μM |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4400 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
10.09 |
-46.19 |
0 |
5 |
-1 |
80 |
437.622 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S)-1-[[(1S,4aS,8aS)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methyl]-5-(3-furyl
[(1S)-1-[[(1S,4aS,8aS)-2,5,5,8a-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-2-E |
Dual Specificity Protein Phosphatase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4600 |
0.25 |
Binding ≤ 10μM |
|
MPIP1-1-E |
Dual Specificity Phosphatase Cdc25A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.25 |
Binding ≤ 10μM |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4400 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
10.31 |
-50.75 |
0 |
5 |
-1 |
80 |
437.622 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S,2S)-1-[[(1S,4aS,8aS)-2,5,5,8a-tetramethyl-1,4,4a,6,7,8-hexahydronaphthalen-1-yl]methyl]-8-(2,
[(E,1S,2S)-1-[[(1S,4aS,8aS)-2,5,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.33 |
9.53 |
-50.75 |
2 |
6 |
-1 |
107 |
547.778 |
11 |
↓
|
|