|
|
Analogs
-
968376
-
-
3830821
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
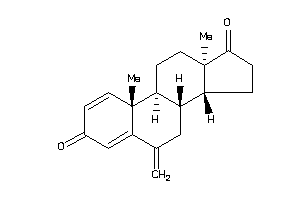
(8S,9S,10R,13R,14S)-10,13-dimethyl-6-methylene-7,8,9,11,12,14,15,16-octahydrocyclopenta[a]phenanthre
(8S,9S,10R,13R,14S)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
9.5 |
-11.96 |
0 |
2 |
0 |
34 |
296.41 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8S,9R,10S,13R,14S,17R)-17-acetyl-10,13-dimethyl-6-methylene-3-oxo-1,2,7,8,9,11,12,14,15,16-decahyd
[(8S,9R,10S,13R,14S,17R)-17-acet…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.34 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
11.63 |
-15.89 |
0 |
4 |
0 |
60 |
384.516 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8S,9R,10S,13R,14R,17R)-17-acetyl-10,13-dimethyl-6-methylene-3-oxo-1,2,7,8,9,11,12,14,15,16-decahyd
[(8S,9R,10S,13R,14R,17R)-17-acet…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.34 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
11.4 |
-12.82 |
0 |
4 |
0 |
60 |
384.516 |
3 |
↓
|
|
|
|
Analogs
-
3830353
-
-
3830354
-
-
3830355
-
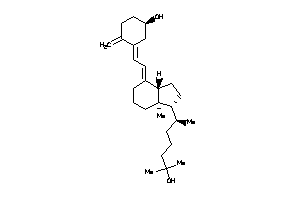
-
3830356
-
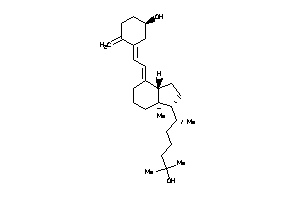
-
3947571
-
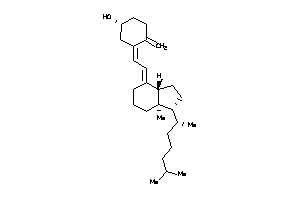
Draw
Identity
99%
90%
80%
70%
Popular Name:
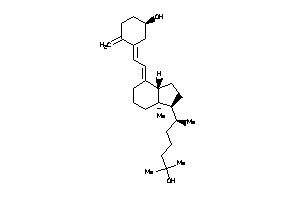
(1R,3Z)-3-[(2E)-2-[(1S,3aS,7aS)-1-[(1R)-1,5-dimethylhexyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden
(1R,3Z)-3-[(2E)-2-[(1S,3aS,7aS)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
13.63 |
-2.2 |
1 |
1 |
0 |
20 |
384.648 |
6 |
↓
|
|
|
|
Analogs
-
3830353
-

-
3830354
-

-
3830355
-

-
3830356
-

-
3947571
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
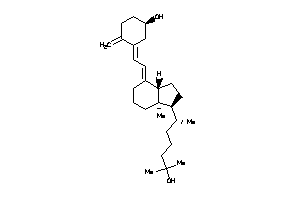
(1R,3Z)-3-[(2E)-2-[(1S,3aS,7aS)-1-[(1S)-1,5-dimethylhexyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden
(1R,3Z)-3-[(2E)-2-[(1S,3aS,7aS)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
13.57 |
-2.13 |
1 |
1 |
0 |
20 |
384.648 |
6 |
↓
|
|
|
|
Analogs
-
3830353
-

-
3830354
-

-
3830355
-

-
3830356
-

-
3947571
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3Z)-3-[(2E)-2-[(1R,3aS,7aS)-1-[(1R)-1,5-dimethylhexyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden
(1R,3Z)-3-[(2E)-2-[(1R,3aS,7aS)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
13.7 |
-2.33 |
1 |
1 |
0 |
20 |
384.648 |
6 |
↓
|
|
|
|
Analogs
-
3830353
-

-
3830354
-

-
3830355
-

-
3830356
-

-
3947571
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3Z)-3-[(2E)-2-[(1R,3aS,7aS)-1-[(1S)-1,5-dimethylhexyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden
(1R,3Z)-3-[(2E)-2-[(1R,3aS,7aS)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
13.64 |
-2.31 |
1 |
1 |
0 |
20 |
384.648 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3E)-3-[(2E)-2-[(1S,3aS,7aR)-1-[(E,1R)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1
(1R,3E)-3-[(2E)-2-[(1S,3aS,7aR)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
13.51 |
-2.16 |
1 |
1 |
0 |
20 |
382.632 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3E)-3-[(2E)-2-[(1S,3aS,7aR)-1-[(E,1S)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1
(1R,3E)-3-[(2E)-2-[(1S,3aS,7aR)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
13.31 |
-2.41 |
1 |
1 |
0 |
20 |
382.632 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3E)-3-[(2E)-2-[(1R,3aS,7aR)-1-[(E,1R)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1
(1R,3E)-3-[(2E)-2-[(1R,3aS,7aR)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
13.67 |
-2.22 |
1 |
1 |
0 |
20 |
382.632 |
5 |
↓
|
|
|
|
Analogs
-
38791855
-
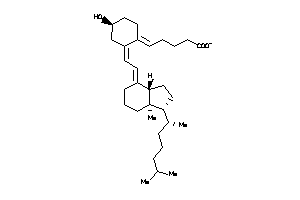
-
38791856
-
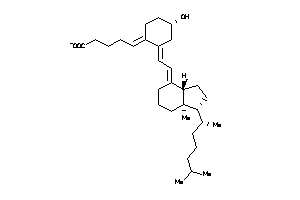
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3E)-3-[(2E)-2-[(1R,3aS,7aR)-1-[(E,1S)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1
(1R,3E)-3-[(2E)-2-[(1R,3aS,7aR)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
13.06 |
-2.18 |
1 |
1 |
0 |
20 |
382.632 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3E,5S)-3-[(2E)-2-[(1S,3aS,7aR)-1-[(E,1R)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahyd
[(1S,3E,5S)-3-[(2E)-2-[(1S,3aS,7…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.72 |
14.59 |
-6.31 |
1 |
3 |
0 |
47 |
440.668 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3E,5S)-3-[(2E)-2-[(1S,3aS,7aR)-1-[(E,1S)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahyd
[(1S,3E,5S)-3-[(2E)-2-[(1S,3aS,7…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.72 |
14.39 |
-6.73 |
1 |
3 |
0 |
47 |
440.668 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3E,5S)-3-[(2E)-2-[(1R,3aS,7aR)-1-[(E,1R)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahyd
[(1S,3E,5S)-3-[(2E)-2-[(1R,3aS,7…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.72 |
14.75 |
-6.43 |
1 |
3 |
0 |
47 |
440.668 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,3E,5S)-3-[(2E)-2-[(1R,3aS,7aR)-1-[(E,1S)-1,5-dimethylhex-2-enyl]-7a-methyl-2,3,3a,5,6,7-hexahyd
[(1S,3E,5S)-3-[(2E)-2-[(1R,3aS,7…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.72 |
14.15 |
-6.36 |
1 |
3 |
0 |
47 |
440.668 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3S,5Z)-5-[(2E)-2-[(3aS,7R,7aS)-7-(6-hydroxy-6-methyl-heptyl)-7a-methyl-2,3,3a,5,6,7-hexahydro-1H
(1R,3S,5Z)-5-[(2E)-2-[(3aS,7R,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.82 |
7.53 |
-5.73 |
3 |
3 |
0 |
61 |
416.646 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.33 |
8.34 |
-5.65 |
3 |
3 |
0 |
61 |
430.673 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3S,5Z)-5-[(2E)-2-[(3aS,7R,7aS)-7-(8-hydroxy-8-methyl-nonyl)-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-
(1R,3S,5Z)-5-[(2E)-2-[(3aS,7R,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.83 |
9.1 |
-5.71 |
3 |
3 |
0 |
61 |
444.7 |
9 |
↓
|
|
|
|
Analogs
-
33820648
-
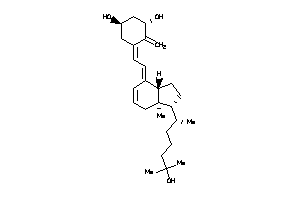
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.51 |
10.78 |
-3.94 |
2 |
2 |
0 |
40 |
412.658 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,5E)-5-[(2E)-2-[(1R,3aR,7aR)-1-[(E,1S,4R)-4-cyclopropyl-4-hydroxy-1-methyl-but-2-enyl]-7a-meth
(1S,3S,5E)-5-[(2E)-2-[(1R,3aR,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
7.49 |
-6.12 |
3 |
3 |
0 |
61 |
412.614 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3S,5E)-5-[(2E)-2-[(1R,3aR,7aR)-1-[(E,1S,4S)-4-cyclopropyl-4-hydroxy-1-methyl-but-2-enyl]-7a-meth
(1S,3S,5E)-5-[(2E)-2-[(1R,3aR,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
7.97 |
-5.5 |
3 |
3 |
0 |
61 |
412.614 |
5 |
↓
|
|
|
|
Analogs
-
38791568
-
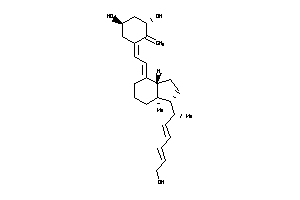
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5Z,7E,22E,24E)-24a,26a,27a-Trihomo-9,10-secocholesta-5,7,10(19),22,24-pentaene-1alpha,3beta,25-triol; 1(S),3(R)-dihydroxy-20(R)-(5'-ethyl-5'-hydroxyhepta-1'(E),3'(E)-dien-1'-yl)-9,10-secopregna-5(Z),7(E),10(19)-triene; 1,3-Cyclohexanediol, 5-((1-(6-ethyl
(5Z,7E,22E,24E)-24a,26a,27a-Trih…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
9.4 |
-5.51 |
3 |
3 |
0 |
61 |
454.695 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3Z,5S)-3-[(2E)-2-[(3aR,7aS)-1-[(E,1S)-5-ethyl-5-hydroxy-1-methyl-hept-3-enyl]-7a-methyl-3a,5,6,7
(1R,3Z,5S)-3-[(2E)-2-[(3aR,7aS)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.05 |
12.49 |
-5.08 |
2 |
2 |
0 |
40 |
442.659 |
7 |
↓
|
|
|
|
Analogs
-
38791653
-
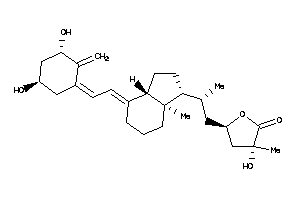
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S)-5-[(2R)-2-[(1S,3aS,4E,7aR)-4-[(2Z)-2-[(3S,5R)-3,5-dihydroxy-2-methylene-cyclohexylidene]ethy
(3R,5S)-5-[(2R)-2-[(1S,3aS,4E,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
5.93 |
-10.35 |
3 |
5 |
0 |
87 |
444.612 |
4 |
↓
|
|
|
|
Analogs
-
38791653
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S)-5-[(2R)-2-[(1S,3aR,4E,7aR)-4-[(2Z)-2-[(3S,5R)-3,5-dihydroxy-2-methylene-cyclohexylidene]ethy
(3R,5S)-5-[(2R)-2-[(1S,3aR,4E,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
6.13 |
-10.64 |
3 |
5 |
0 |
87 |
444.612 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S)-5-[(2R)-2-[(1R,3aR,4E,7aR)-4-[(2Z)-2-[(3S,5R)-3,5-dihydroxy-2-methylene-cyclohexylidene]ethy
(3R,5S)-5-[(2R)-2-[(1R,3aR,4E,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
6.09 |
-10.97 |
3 |
5 |
0 |
87 |
444.612 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.47 |
10.28 |
-9.39 |
3 |
5 |
0 |
87 |
512.731 |
10 |
↓
|
|
|
|
Analogs
-
3921872
-
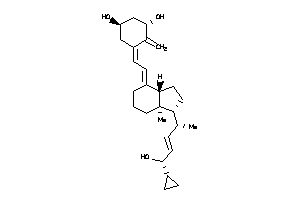
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aS)-1-[(E,1R,4S)-4-cyclopropyl-4-hydroxy-1-methyl-but-2-enyl]-7a-meth
(1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
7.75 |
-5.22 |
3 |
3 |
0 |
61 |
412.614 |
5 |
↓
|
|
|
|
Analogs
-
3830821
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
9.43 |
-16.17 |
0 |
2 |
0 |
34 |
296.41 |
0 |
↓
|
|
|
|
Analogs
-
3830821
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
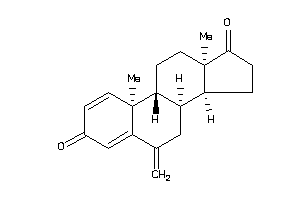
(8R,9S,10R,13S,14R)-10,13-dimethyl-6-methylene-7,8,9,11,12,14,15,16-octahydrocyclopenta[a]phenanthre
(8R,9S,10R,13S,14R)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
9.75 |
-11.92 |
0 |
2 |
0 |
34 |
296.41 |
0 |
↓
|
|