|
|
Analogs
-
11979498
-
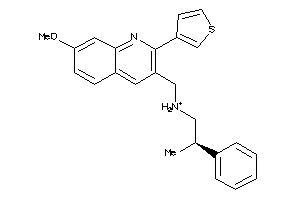
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.54 |
-0.05 |
-61.07 |
2 |
3 |
1 |
39 |
389.544 |
7 |
↓
|
|
|
|
Analogs
-
11979487
-
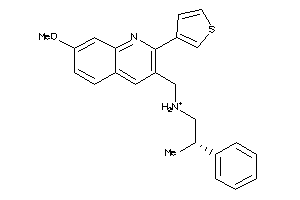
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.54 |
-0.27 |
-50.74 |
2 |
3 |
1 |
39 |
389.544 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
8.54 |
-10.61 |
2 |
4 |
0 |
54 |
469.404 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.84 |
9.76 |
-65.09 |
3 |
4 |
1 |
59 |
470.412 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
10.59 |
-10.93 |
1 |
4 |
0 |
43 |
483.431 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.14 |
11.82 |
-65.57 |
2 |
4 |
1 |
48 |
484.439 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
5000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.95 |
12.01 |
-9.03 |
1 |
2 |
0 |
25 |
437.406 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.95 |
13.24 |
-61.14 |
2 |
2 |
1 |
29 |
438.414 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.95 |
13.34 |
-105.41 |
3 |
2 |
2 |
31 |
439.422 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6900 |
0.27 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
6900 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.17 |
11.54 |
-8.54 |
1 |
2 |
0 |
25 |
457.824 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.17 |
13.08 |
-64.8 |
2 |
2 |
1 |
29 |
458.832 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.93 |
11.59 |
-11.21 |
1 |
3 |
0 |
34 |
467.432 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.93 |
12.82 |
-65.93 |
2 |
3 |
1 |
39 |
468.44 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.47 |
10.83 |
-8.49 |
1 |
3 |
0 |
34 |
507.375 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.47 |
12.36 |
-65.68 |
2 |
3 |
1 |
39 |
508.383 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.41 |
11.58 |
-57.08 |
1 |
4 |
-1 |
65 |
466.38 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.41 |
13.13 |
-104.85 |
2 |
4 |
0 |
70 |
467.388 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5100 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.02 |
8.2 |
-10.5 |
2 |
3 |
0 |
45 |
439.378 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.02 |
9.72 |
-64.94 |
3 |
3 |
1 |
50 |
440.386 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
8.54 |
-10.84 |
2 |
4 |
0 |
54 |
469.404 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.84 |
9.76 |
-66.11 |
3 |
4 |
1 |
59 |
470.412 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
6.15 |
-11.52 |
3 |
4 |
0 |
65 |
455.377 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.53 |
7.69 |
-66.36 |
4 |
4 |
1 |
70 |
456.385 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.55 |
10.65 |
-8.74 |
1 |
3 |
0 |
34 |
453.405 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.55 |
11.88 |
-59.72 |
2 |
3 |
1 |
39 |
454.413 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.02 |
8.18 |
-9.03 |
2 |
3 |
0 |
45 |
439.378 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.02 |
9.71 |
-62.05 |
3 |
3 |
1 |
50 |
440.386 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
65 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.55 |
10.67 |
-11.35 |
1 |
3 |
0 |
34 |
453.405 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.55 |
11.89 |
-66.01 |
2 |
3 |
1 |
39 |
454.413 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
2800 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.53 |
10.65 |
-11.42 |
1 |
3 |
0 |
34 |
453.405 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.53 |
11.88 |
-64.73 |
2 |
3 |
1 |
39 |
454.413 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.28 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR_HUMAN |
Q8TDU6
|
G-protein Coupled Bile Acid Receptor 1, Human |
5500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
11.36 |
-8.87 |
1 |
2 |
0 |
25 |
423.379 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.52 |
12.58 |
-60.94 |
2 |
2 |
1 |
29 |
424.387 |
6 |
↓
|
|