|
|
Analogs
-
36143609
-
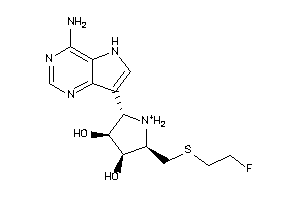
-
26961672
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTAP-1-E |
S-methyl-5-thioadenosine Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
0.1 |
-44.02 |
7 |
7 |
1 |
125 |
328.393 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.79 |
-1 |
-11.15 |
6 |
7 |
0 |
120 |
327.385 |
5 |
↓
|
|
|
|
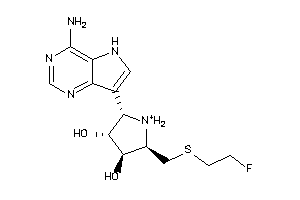
Analogs
-
36143606
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTAP-1-E |
S-methyl-5-thioadenosine Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
-0.31 |
-48.99 |
7 |
7 |
1 |
125 |
328.393 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.79 |
-1.44 |
-16.56 |
6 |
7 |
0 |
120 |
327.385 |
5 |
↓
|
|
|
|
|
|
|
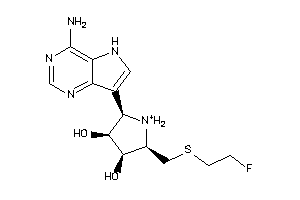
Analogs
-
36143613
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTAP-1-E |
S-methyl-5-thioadenosine Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.51 |
2.77 |
-42.34 |
7 |
7 |
1 |
125 |
372.474 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.51 |
1.6 |
-12.44 |
6 |
7 |
0 |
120 |
371.466 |
5 |
↓
|
|
|
|
|
|
|
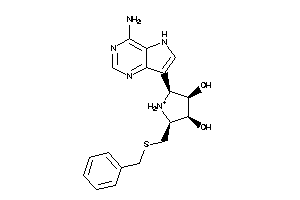
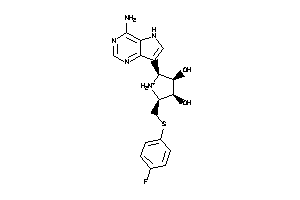
Analogs
-
36143620
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTAP-1-E |
S-methyl-5-thioadenosine Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.61 |
1.93 |
-47.55 |
7 |
7 |
1 |
125 |
376.437 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.61 |
0.77 |
-12.69 |
6 |
7 |
0 |
120 |
375.429 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
3960046
-
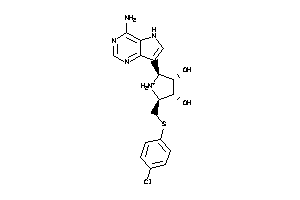
-
36143626
-
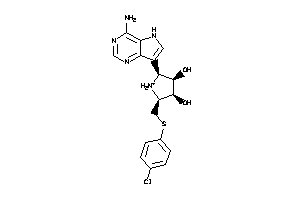
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTAP-1-E |
S-methyl-5-thioadenosine Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
2.38 |
-46.76 |
7 |
7 |
1 |
125 |
392.892 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.12 |
1.22 |
-12.28 |
6 |
7 |
0 |
120 |
391.884 |
4 |
↓
|
|
|
|
Analogs
-
13647999
-
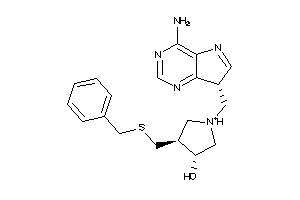
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTAP-1-E |
S-methyl-5-thioadenosine Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
7.21 |
-44.76 |
5 |
6 |
1 |
92 |
370.502 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.60 |
4.55 |
-11.79 |
4 |
6 |
0 |
91 |
369.494 |
6 |
↓
|
|
|
|
Analogs
-
38245886
-
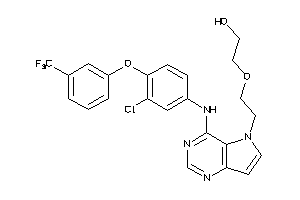
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
8.16 |
-13.82 |
2 |
8 |
0 |
94 |
493.873 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.61 |
14 |
-9.71 |
0 |
4 |
0 |
34 |
376.548 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
5.69 |
-11.35 |
0 |
5 |
0 |
65 |
307.762 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.90 |
1.92 |
-10.85 |
3 |
4 |
0 |
68 |
213.038 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
2.96 |
-27.47 |
0 |
4 |
-1 |
64 |
177.574 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.04 |
3.41 |
-13.07 |
1 |
4 |
0 |
65 |
178.582 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
3.01 |
-9.29 |
1 |
3 |
0 |
42 |
171.562 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.45 |
2.55 |
-30.04 |
0 |
3 |
-1 |
40 |
170.554 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
2.72 |
-8.74 |
1 |
4 |
0 |
59 |
181.582 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.07 |
2.25 |
-28.95 |
0 |
4 |
-1 |
57 |
180.574 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
3.42 |
-4.25 |
1 |
3 |
0 |
42 |
232.468 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
2.97 |
-28.91 |
0 |
3 |
-1 |
40 |
231.46 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
3.38 |
-8.68 |
1 |
3 |
0 |
42 |
266.913 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.09 |
2.92 |
-26.41 |
0 |
3 |
-1 |
40 |
265.905 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
3.17 |
-4.15 |
1 |
3 |
0 |
42 |
266.913 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
2.73 |
-26.44 |
0 |
3 |
-1 |
40 |
265.905 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
2.91 |
-8.68 |
3 |
4 |
0 |
68 |
247.483 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.90 |
2.46 |
-30.34 |
2 |
4 |
-1 |
66 |
246.475 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
2.71 |
-4.96 |
3 |
4 |
0 |
68 |
247.483 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.09 |
2.26 |
-30.84 |
2 |
4 |
-1 |
66 |
246.475 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
114 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
6.97 |
-56.85 |
4 |
7 |
1 |
93 |
385.879 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.34 |
6.67 |
-13.81 |
3 |
7 |
0 |
91 |
384.871 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.24 |
-6.86 |
-12.87 |
7 |
8 |
0 |
140 |
265.273 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.24 |
-5.57 |
-43.14 |
8 |
8 |
1 |
145 |
266.281 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
4.56 |
-14.54 |
1 |
5 |
0 |
68 |
225.635 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
4.29 |
-9.5 |
0 |
3 |
0 |
31 |
202.044 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
2.33 |
-11.3 |
1 |
3 |
0 |
42 |
153.572 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
-0.2 |
-43.44 |
1 |
4 |
-1 |
65 |
213.014 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
0.74 |
1.83 |
-12.45 |
2 |
4 |
0 |
62 |
214.022 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.74 |
1.16 |
-17.3 |
2 |
4 |
0 |
62 |
214.022 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.34 |
10.44 |
-124.46 |
6 |
11 |
-1 |
193 |
453.479 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.34 |
8.45 |
-78.83 |
7 |
11 |
0 |
191 |
454.487 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.34 |
10.53 |
-124.22 |
6 |
11 |
-1 |
193 |
453.479 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.34 |
10.07 |
-124.14 |
5 |
11 |
-2 |
192 |
452.471 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.34 |
8.55 |
-77.67 |
7 |
11 |
0 |
191 |
454.487 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.34 |
10.49 |
-123.95 |
6 |
11 |
-1 |
193 |
453.479 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.34 |
10.03 |
-125.74 |
5 |
11 |
-2 |
192 |
452.471 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.34 |
8.52 |
-76.42 |
7 |
11 |
0 |
191 |
454.487 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.89 |
10.12 |
-124.95 |
6 |
11 |
-1 |
193 |
439.452 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.89 |
9.72 |
-125.93 |
5 |
11 |
-2 |
192 |
438.444 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.89 |
8.14 |
-78.9 |
7 |
11 |
0 |
191 |
440.46 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-1.62 |
-45.74 |
6 |
7 |
1 |
112 |
264.309 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.15 |
-4.16 |
-17.7 |
5 |
7 |
0 |
111 |
263.301 |
3 |
↓
|
|