|
|
|
|
|
|
|
|
Analogs
-
36160097
-
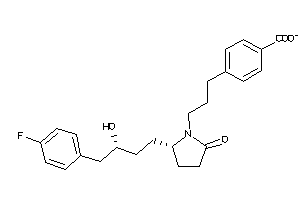
-
36160081
-
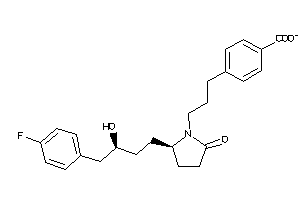
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2767 |
0.26 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R4_RAT |
P43114
|
Prostanoid EP4 Receptor, Rat |
2767 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
11.83 |
-61.36 |
1 |
5 |
-1 |
81 |
412.481 |
10 |
↓
|
|
|
|
Analogs
-
36160081
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2767 |
0.26 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R4_RAT |
P43114
|
Prostanoid EP4 Receptor, Rat |
2767 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
11.65 |
-64.78 |
1 |
5 |
-1 |
81 |
412.481 |
10 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
36160143
-
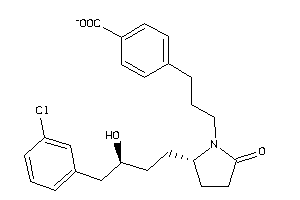
-
36160126
-
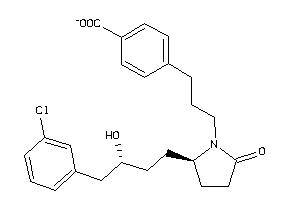
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
42 |
0.34 |
Binding ≤ 10μM
|
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
425 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.64 |
12.19 |
-61.72 |
1 |
5 |
-1 |
81 |
428.936 |
10 |
↓
|
|
|
|
Analogs
-
36160126
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
42 |
0.34 |
Binding ≤ 10μM
|
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
425 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.64 |
12.22 |
-60.98 |
1 |
5 |
-1 |
81 |
428.936 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.30 |
Binding ≤ 10μM
|
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
105 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.86 |
12.67 |
-62.8 |
1 |
5 |
-1 |
81 |
462.488 |
11 |
↓
|
|