|
|
Analogs
-
42852815
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
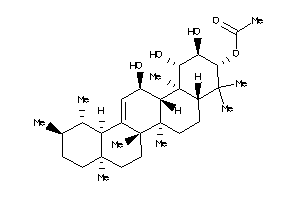
[(1S,2R,3R,4aS,6aR,6bS,8aR,12aR,14R,14aS,14bS)-1,2,14-trihydroxy-4,4,6a,6b,8a,11,11,14b-octamethyl-1
[(1S,2R,3R,4aS,6aR,6bS,8aR,12aR,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.20 |
9.75 |
-8.31 |
3 |
5 |
0 |
87 |
516.763 |
2 |
↓
|
|
|
|
Analogs
-
3978269
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
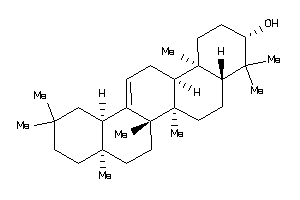
(3R,4aR,6aR,6bR,8aS,11S,12R,12aS,14aS,14bS)-4,4,6a,6b,8a,11,12,14b-octamethyl-2,3,4a,5,6,7,8,9,10,11
(3R,4aR,6aR,6bR,8aS,11S,12R,12aS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.08 |
12.49 |
-1.28 |
1 |
1 |
0 |
20 |
426.729 |
0 |
↓
|
|
|
|
Analogs
-
3978269
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,4aS,6aR,6bR,8aS,12aR,14aR,14bS)-4,4,6a,6b,8a,11,11,14b-octamethyl-1,2,3,4a,5,6,7,8,9,10,12,12a,1
(3R,4aS,6aR,6bR,8aS,12aR,14aR,14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.02 |
12.52 |
-1.15 |
1 |
1 |
0 |
20 |
426.729 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
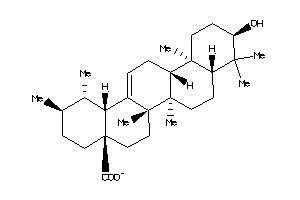
(4aS,6aR,6aS,6bR,8aR,9S,10R,12aR,14bS)-10-hydroxy-9-(hydroxymethyl)-2,2,6a,6b,9,12a-hexamethyl-1,3,4
(4aS,6aR,6aS,6bR,8aR,9S,10R,12aR…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.55 |
9.24 |
-54.84 |
2 |
4 |
-1 |
81 |
471.702 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.55 |
7.28 |
-7.72 |
3 |
4 |
0 |
78 |
472.71 |
2 |
↓
|
|
|
|
Analogs
-
2117641
-
-
3941105
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
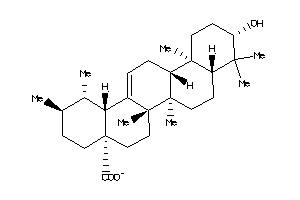
(1R,2S,4aR,6aR,6aS,6bS,8aR,10R,12aS,14bR)-10-hydroxy-1,2,6a,6b,9,9,12a-heptamethyl-2,3,4,5,6,6a,7,8,
(1R,2S,4aR,6aR,6aS,6bS,8aR,10R,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
12.28 |
-55 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.79 |
10.34 |
-6.35 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
-
34953949
-
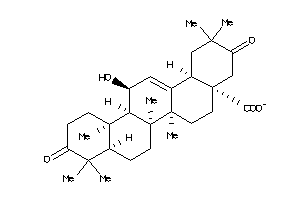
-
34953950
-
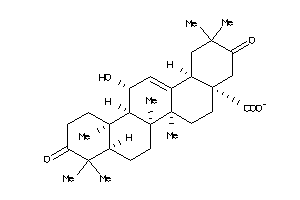
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aS,6aR,6bR,8aR,10S,12aS,13S,14bS)-10,13-dihydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,
(2S,4aS,6aS,6aR,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
8.96 |
-52.51 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
6.99 |
-7.45 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|
|
|
Analogs
-
34953949
-

-
34953950
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aR,6aR,6bR,8aR,10S,12aS,13S,14bS)-10,13-dihydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,
(2S,4aS,6aR,6aR,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
9.62 |
-49.71 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
7.66 |
-6.23 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|
|
|
Analogs
-
34953949
-

-
34953950
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aS,6aR,6bR,8aR,10S,12aS,13R,14bS)-10,13-dihydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,
(2S,4aS,6aS,6aR,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
9.71 |
-50.74 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
7.73 |
-6.5 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|
|
|
Analogs
-
34953949
-

-
34953950
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aR,6aR,6bR,8aR,10S,12aS,13R,14bS)-10,13-dihydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,
(2S,4aS,6aR,6aR,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
9.38 |
-52.01 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
7.42 |
-6.93 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|
|
|
Analogs
-
40916316
-
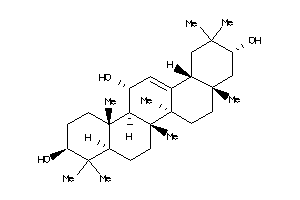
-
40919175
-
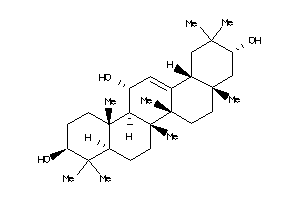
-
4102266
-
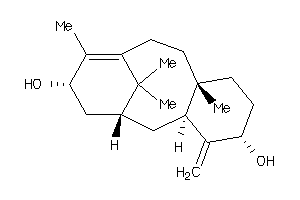
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4aR,6aR,6bR,8aS,11S,12aS,14S,14aS,14bS)-11-(hydroxymethyl)-4,4,6a,6b,8a,11,14b-heptamethyl-1,2,3
(3S,4aR,6aR,6bR,8aS,11S,12aS,14S…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.93 |
5.89 |
-4.35 |
3 |
3 |
0 |
61 |
458.727 |
1 |
↓
|
|
|
|
Analogs
-
4102266
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4aR,6aR,6bR,8aS,11S,12aS,14S,14aR,14bS)-11-(hydroxymethyl)-4,4,6a,6b,8a,11,14b-heptamethyl-1,2,3
(3S,4aR,6aR,6bR,8aS,11S,12aS,14S…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.93 |
6.51 |
-4.25 |
3 |
3 |
0 |
61 |
458.727 |
1 |
↓
|
|
|
|
Analogs
-
5762239
-
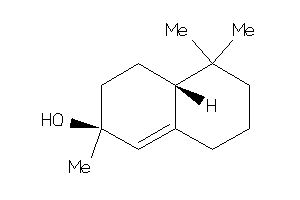
-
5762240
-
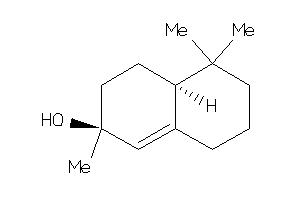
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4aR,6aR,6bR,8aS,11S,12aS,14R,14aS,14bS)-11-(hydroxymethyl)-4,4,6a,6b,8a,11,14b-heptamethyl-1,2,3
(3S,4aR,6aR,6bR,8aS,11S,12aS,14R…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.93 |
6.56 |
-4.35 |
3 |
3 |
0 |
61 |
458.727 |
1 |
↓
|
|
|
|
Analogs
-
4102266
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4aR,6aR,6bR,8aS,11S,12aS,14R,14aR,14bS)-11-(hydroxymethyl)-4,4,6a,6b,8a,11,14b-heptamethyl-1,2,3
(3S,4aR,6aR,6bR,8aS,11S,12aS,14R…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.93 |
6.24 |
-4.66 |
3 |
3 |
0 |
61 |
458.727 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,2R,4aR,6aS,6aR,6bR,8aS,10S,11S,12aR,14bS)-10,11-dihydroxy-1,2,6a,6b,9,9,12a-heptamethyl-2,3,4,5,
(1S,2R,4aR,6aS,6aR,6bR,8aS,10S,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
810 |
0.25 |
Binding ≤ 10μM
|
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5490 |
0.22 |
Binding ≤ 10μM
|
|
Z80874-6-O |
CEM (T-cell Leukemia) (cluster #6 Of 7), Other |
Other |
3270 |
0.23 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.87 |
9.77 |
-53.8 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.87 |
7.83 |
-7.4 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
10.16 |
27.35 |
-3.85 |
0 |
2 |
0 |
26 |
665.144 |
16 |
↓
|
|
|
|
|
|
|
Analogs
-
8035519
-
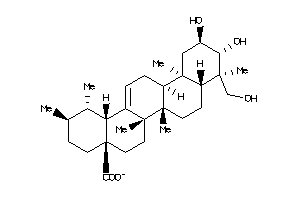
-
8221271
-
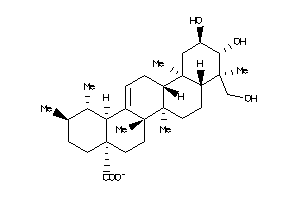
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
5.48 |
-58.46 |
3 |
5 |
-1 |
101 |
487.701 |
2 |
↓
|
|
|
|
Analogs
-
8035519
-

-
8221271
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
6.98 |
-58.59 |
3 |
5 |
-1 |
101 |
487.701 |
2 |
↓
|
|
|
|
Analogs
-
8035519
-

-
8221271
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
4.38 |
-59 |
4 |
6 |
-1 |
121 |
503.7 |
2 |
↓
|
|
|
|
Analogs
-
8035519
-

-
8221271
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
4.47 |
-67.42 |
4 |
6 |
-1 |
121 |
503.7 |
2 |
↓
|
|
|
|
Analogs
-
8035519
-

-
8221271
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
2.41 |
-61.37 |
4 |
6 |
-1 |
121 |
503.7 |
2 |
↓
|
|
|
|
Analogs
-
8035519
-

-
8221271
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
4.29 |
-58.19 |
4 |
6 |
-1 |
121 |
503.7 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aR,6aS,6aR,6bR,8aS,9R,10R,11S,12aR,14bR)-10,11-dihydroxy-9-(hydroxymethyl)-2,6a,6b,9,12a-pentam
(2S,4aR,6aS,6aR,6bR,8aS,9R,10R,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
6.05 |
-129.57 |
3 |
7 |
-2 |
141 |
516.675 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
4.23 |
-63.71 |
4 |
7 |
-1 |
138 |
517.683 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
4.08 |
-55.6 |
4 |
7 |
-1 |
138 |
517.683 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4S,4aR,6aR,6bS,8aS,11R,12R,12aS,14aS,14bR)-3,12-dihydroxy-4,6a,6b,11,12,14b-hexamethyl-1,2,3,4a,
(3S,4S,4aR,6aR,6bS,8aS,11R,12R,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
9.9 |
-111.29 |
2 |
6 |
-2 |
121 |
500.676 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.55 |
7.93 |
-62.53 |
3 |
6 |
-1 |
118 |
501.684 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4S,4aR,6aR,6bS,8aS,11R,12R,12aS,14aR,14bR)-3,12-dihydroxy-4,6a,6b,11,12,14b-hexamethyl-1,2,3,4a,
(3S,4S,4aR,6aR,6bS,8aS,11R,12R,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
9.81 |
-109.6 |
2 |
6 |
-2 |
121 |
500.676 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.55 |
8.11 |
-58.32 |
3 |
6 |
-1 |
118 |
501.684 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4S,4aS,6aR,6bS,8aS,11R,12R,12aS,14aS,14bR)-3,12-dihydroxy-4,6a,6b,11,12,14b-hexamethyl-1,2,3,4a,
(3S,4S,4aS,6aR,6bS,8aS,11R,12R,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
9.83 |
-105.55 |
2 |
6 |
-2 |
121 |
500.676 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.55 |
7.98 |
-57.22 |
3 |
6 |
-1 |
118 |
501.684 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4S,4aS,6aR,6bS,8aS,11R,12R,12aS,14aR,14bR)-3,12-dihydroxy-4,6a,6b,11,12,14b-hexamethyl-1,2,3,4a,
(3S,4S,4aS,6aR,6bS,8aS,11R,12R,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
9.61 |
-102.19 |
2 |
6 |
-2 |
121 |
500.676 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.55 |
7.89 |
-44.88 |
3 |
6 |
-1 |
118 |
501.684 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aR,6aR,6aS,6bR,8aR,10S,12aR,14bR)-10-hydroxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,
(4aR,6aR,6aS,6bR,8aR,10S,12aR,14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.73 |
12.1 |
-50.83 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.73 |
10.24 |
-5.72 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,6aR,6aR,6bR,8aR,10R,12aS,14bR)-10-acetoxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,
(4aS,6aR,6aR,6bR,8aR,10R,12aS,14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.43 |
16.55 |
-49.66 |
0 |
4 |
-1 |
66 |
497.74 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.43 |
14.92 |
-7.74 |
1 |
4 |
0 |
64 |
498.748 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,6aR,6aR,6bR,8aR,10S,12aS,14bR)-10-acetoxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,
(4aS,6aR,6aR,6bR,8aR,10S,12aS,14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.43 |
16.53 |
-47.61 |
0 |
4 |
-1 |
66 |
497.74 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.43 |
14.91 |
-7.21 |
1 |
4 |
0 |
64 |
498.748 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,6aR,6aR,6bR,8aR,10R,12aR,14bR)-10-acetoxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,
(4aS,6aR,6aR,6bR,8aR,10R,12aR,14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.43 |
16.4 |
-48.16 |
0 |
4 |
-1 |
66 |
497.74 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.43 |
14.79 |
-7.55 |
1 |
4 |
0 |
64 |
498.748 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,6aR,6aR,6bR,8aR,10S,12aR,14bR)-10-acetoxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,
(4aS,6aR,6aR,6bR,8aR,10S,12aR,14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.43 |
16.43 |
-52.59 |
0 |
4 |
-1 |
66 |
497.74 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.43 |
14.48 |
-8.97 |
1 |
4 |
0 |
64 |
498.748 |
3 |
↓
|
|
|
|
Analogs
-
14980264
-
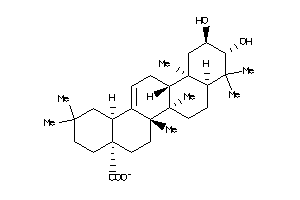
-
14980267
-
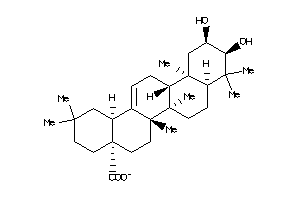
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,6aS,6aS,6bR,8aR,10S,11R,12aR,14bS)-10,11-dihydroxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7
(4aS,6aS,6aS,6bR,8aR,10S,11R,12a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
10.12 |
-59.5 |
2 |
4 |
-1 |
81 |
471.702 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
8.17 |
-9.47 |
3 |
4 |
0 |
78 |
472.71 |
1 |
↓
|
|