|
|
|
|
|
Analogs
-
36215476
-
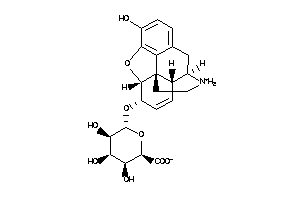
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6S)-3,4,5-trihydroxy-6-(hydroxyBLAHyl)oxy-tetrahydropyran-2-carboxylic
(2R,3S,4R,5R,6S)-3,4,5-trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
0.59 |
-101.76 |
6 |
10 |
0 |
165 |
447.44 |
3 |
↓
|
|
|
|
Analogs
-
36221452
-
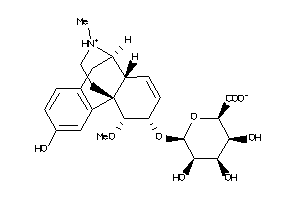
-
36221454
-
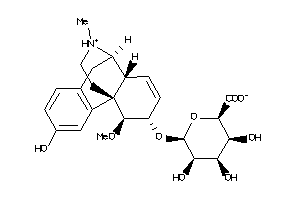
-
36215473
-
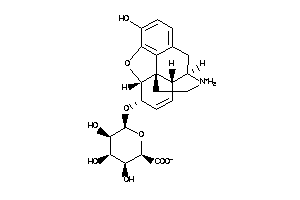
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6R)-3,4,5-trihydroxy-6-(hydroxyBLAHyl)oxy-tetrahydropyran-2-carboxylic
(2R,3S,4R,5R,6R)-3,4,5-trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-0.6 |
-84.16 |
6 |
10 |
0 |
165 |
447.44 |
3 |
↓
|
|
|
|
Analogs
-
36215489
-
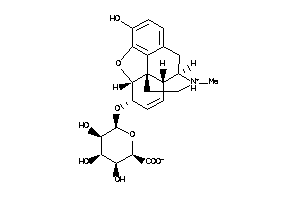
-
36215499
-
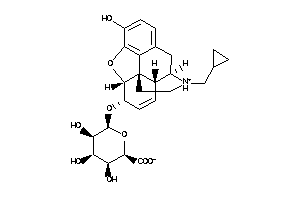
-
36215506
-
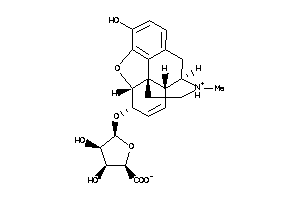
-
36221452
-

-
36221454
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6S)-3,4,5-trihydroxy-6-[methoxy(methyl)BLAHyl]oxy-tetrahydropyran-2-carboxylic
(2R,3S,4R,5R,6S)-3,4,5-trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
85 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.42 |
4.79 |
-95.87 |
4 |
10 |
0 |
142 |
475.494 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
168 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
8.94 |
-93.46 |
2 |
7 |
0 |
92 |
413.47 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6S)-6-[allyl(hydroxy)BLAHyl]oxy-3,4,5-trihydroxy-tetrahydropyran-2-carboxylic
(2R,3S,4R,5R,6S)-6-[allyl(hydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.08 |
3.01 |
-90.08 |
5 |
10 |
0 |
153 |
487.505 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.08 |
1.64 |
-58.41 |
4 |
10 |
-1 |
152 |
486.497 |
5 |
↓
|
|
|
|
Analogs
-
36215499
-

-
36215506
-

-
36221452
-

-
36221454
-

-
43888391
-
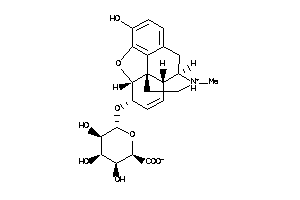
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6S)-3,4,5-trihydroxy-6-[hydroxy(methyl)BLAHyl]oxy-tetrahydropyran-2-carboxylic
(2R,3S,4R,5R,6S)-3,4,5-trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
190 |
0.29 |
Binding ≤ 10μM
|
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.32 |
Binding ≤ 10μM
|
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
72 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_PIG |
P79291
|
Delta Opioid Receptor, Pig |
190 |
0.29 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
190 |
0.29 |
Binding ≤ 1μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
72.3 |
0.30 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
72.3 |
0.30 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_PIG |
P79291
|
Delta Opioid Receptor, Pig |
190 |
0.29 |
Binding ≤ 10μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
190 |
0.29 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
72.3 |
0.30 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
72.3 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.73 |
1.79 |
-92.32 |
5 |
10 |
0 |
153 |
461.467 |
3 |
↓
|
|
|
|
Analogs
-
42878694
-
Draw
Identity
99%
90%
80%
70%
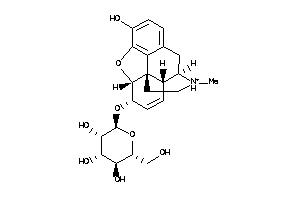
Popular Name:
(2S,3S,4R,5R,6S)-2-(hydroxymethyl)-6-[hydroxy(methyl)BLAHyl]oxy-tetrahydropyran-3,4,5-triol
(2S,3S,4R,5R,6S)-2-(hydroxymethy…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.60 |
-1.65 |
-59.21 |
6 |
9 |
1 |
133 |
448.492 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6S)-6-[carboxymethyl(hydroxy)BLAHyl]oxy-3,4,5-trihydroxy-tetrahydropyran-2-carboxylic
(2R,3S,4R,5R,6S)-6-[carboxymethy…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-4-E |
Mu-type Opioid Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
33 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.39 |
3.19 |
-89.95 |
5 |
12 |
-1 |
193 |
504.468 |
5 |
↓
|
|