|
|
Analogs
-
36215633
-
-
36215637
-
-
36215639
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
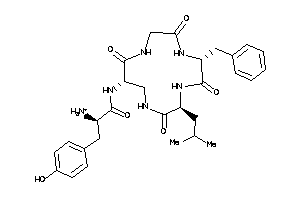
(2R)-2-amino-N-[(1S,7R,10R)-7-benzyl-10-isobutyl-2,5,8,11-tetraoxo-3,6,9,12-tetrazacyclotridec-1-yl]
(2R)-2-amino-N-[(1S,7R,10R)-7-be…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.25 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
96 |
0.24 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
73 |
0.24 |
Functional ≤ 10μM
|
|
OPRM-1-E |
Mu Opioid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
1.36 |
-69.94 |
9 |
12 |
1 |
193 |
567.667 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.19 |
1.04 |
-19.23 |
8 |
12 |
0 |
192 |
566.659 |
8 |
↓
|
|
|
|
Analogs
-
36215637
-

-
36215639
-

-
36215630
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
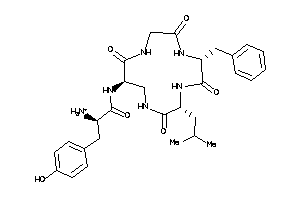
(2R)-2-amino-N-[(1S,7R,10S)-7-benzyl-10-isobutyl-2,5,8,11-tetraoxo-3,6,9,12-tetrazacyclotridec-1-yl]
(2R)-2-amino-N-[(1S,7R,10S)-7-be…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.25 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
96 |
0.24 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
73 |
0.24 |
Functional ≤ 10μM
|
|
OPRM-1-E |
Mu Opioid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
1.52 |
-66.34 |
9 |
12 |
1 |
193 |
567.667 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.19 |
1.2 |
-17.2 |
8 |
12 |
0 |
192 |
566.659 |
8 |
↓
|
|
|
|
Analogs
-
36215639
-

-
36215630
-

-
36215633
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
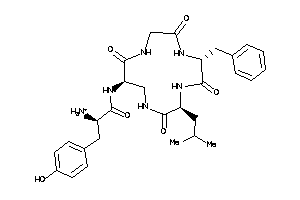
(2R)-2-amino-N-[(1R,7R,10R)-7-benzyl-10-isobutyl-2,5,8,11-tetraoxo-3,6,9,12-tetrazacyclotridec-1-yl]
(2R)-2-amino-N-[(1R,7R,10R)-7-be…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.25 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
96 |
0.24 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
73 |
0.24 |
Functional ≤ 10μM
|
|
OPRM-1-E |
Mu Opioid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
1.25 |
-61.13 |
9 |
12 |
1 |
193 |
567.667 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.19 |
0.93 |
-16.04 |
8 |
12 |
0 |
192 |
566.659 |
8 |
↓
|
|
|
|
Analogs
-
36215630
-

-
36215633
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
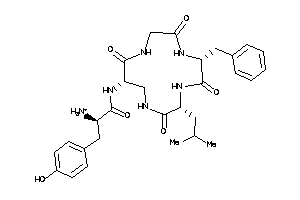
(2R)-2-amino-N-[(1R,7R,10S)-7-benzyl-10-isobutyl-2,5,8,11-tetraoxo-3,6,9,12-tetrazacyclotridec-1-yl]
(2R)-2-amino-N-[(1R,7R,10S)-7-be…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.25 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
96 |
0.24 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
73 |
0.24 |
Functional ≤ 10μM
|
|
OPRM-1-E |
Mu Opioid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
1.41 |
-59.12 |
9 |
12 |
1 |
193 |
567.667 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.19 |
1.09 |
-14.5 |
8 |
12 |
0 |
192 |
566.659 |
8 |
↓
|
|