|
|
Analogs
-
36215665
-
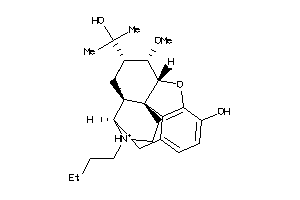
-
36215670
-
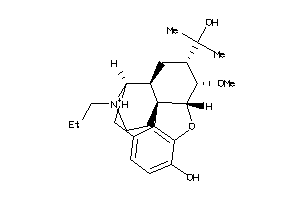
-
38232309
-
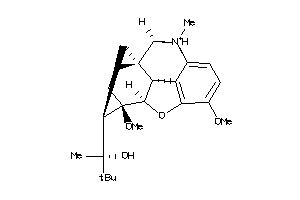
-
38232310
-
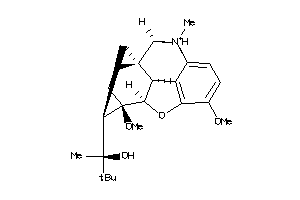
-
60027376
-
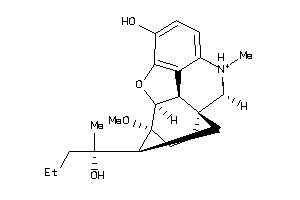
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
OPRX-1-E |
Nociceptin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 1μM
|
|
OPRX_RAT |
P35370
|
Nociceptin Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 10μM
|
|
OPRX_RAT |
P35370
|
Nociceptin Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
8.5 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
6.8 |
-47.2 |
3 |
5 |
1 |
63 |
400.539 |
3 |
↓
|
|
|
|
Analogs
-
6491031
-
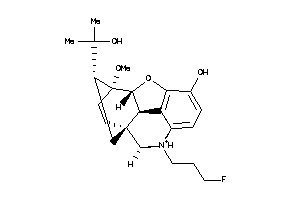
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.72 |
8.28 |
-48.63 |
3 |
5 |
1 |
63 |
446.583 |
5 |
↓
|
|
|
|
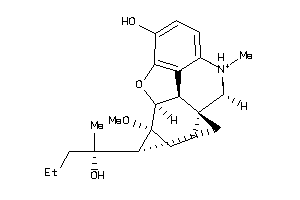
Analogs
-
36215670
-

-
38232309
-

-
38232310
-

-
60027376
-

-
60027377
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
OPRX-1-E |
Nociceptin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 1μM
|
|
OPRX_RAT |
P35370
|
Nociceptin Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 10μM
|
|
OPRX_RAT |
P35370
|
Nociceptin Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
5.9 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
8.33 |
-48.81 |
3 |
5 |
1 |
63 |
428.593 |
5 |
↓
|
|
|
|
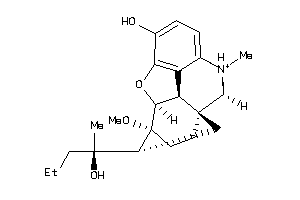
Analogs
-
38232309
-

-
38232310
-

-
60027376
-

-
60027377
-

-
60027378
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
|
OPRX-1-E |
Nociceptin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 1μM
|
|
OPRX_RAT |
P35370
|
Nociceptin Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 10μM
|
|
OPRX_RAT |
P35370
|
Nociceptin Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
2.7 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
7.56 |
-47.79 |
3 |
5 |
1 |
63 |
414.566 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
9.44 |
-53.85 |
3 |
5 |
0 |
63 |
468.658 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6R)-6-[cyclopropylmethyl-[(1R)-1-hydroxy-1,2,2-trimethyl-propyl]-methoxy-BLAHyl]oxy-3,4
(2R,3S,4R,5R,6R)-6-[cyclopropylm…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
5.86 |
-95.15 |
5 |
11 |
0 |
162 |
643.774 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6R)-6-[cyclopropylmethyl-[(1S)-1-hydroxy-1,2,2-trimethyl-propyl]-methoxy-BLAHyl]oxy-3,4
(2R,3S,4R,5R,6R)-6-[cyclopropylm…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
6.52 |
-87.48 |
5 |
11 |
0 |
162 |
643.774 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6R)-6-[cyclopropylmethyl-[(1R)-1-hydroxy-1,2,2-trimethyl-propyl]-methoxy-BLAHyl]oxy-3,4
(2R,3S,4R,5R,6R)-6-[cyclopropylm…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
6.42 |
-94.07 |
5 |
11 |
0 |
162 |
643.774 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4R,5R,6R)-6-[cyclopropylmethyl-[(1S)-1-hydroxy-1,2,2-trimethyl-propyl]-methoxy-BLAHyl]oxy-3,4
(2R,3S,4R,5R,6R)-6-[cyclopropylm…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
6.31 |
-93.45 |
5 |
11 |
0 |
162 |
643.774 |
8 |
↓
|
|