|
|
Analogs
-
8090
-
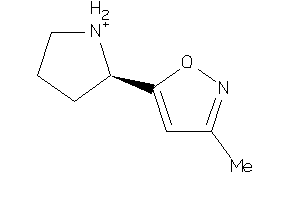
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
3.6 |
-40.56 |
2 |
3 |
1 |
43 |
153.205 |
1 |
↓
|
|
|
|
Analogs
-
4098885
-
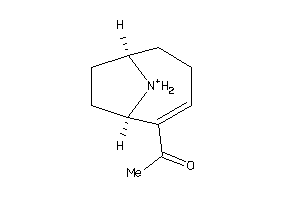
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
90 |
0.82 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB-3-E |
Acetylcholine Receptor Protein Beta Chain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
19 |
0.90 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.94 |
Binding ≤ 10μM
|
|
ACHG-3-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.81 |
Functional ≤ 10μM
|
|
Z104287-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 3), Other |
Other |
25 |
0.89 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
1 |
1.05 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
48 |
0.85 |
Binding ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2000 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
4.05 |
-42.61 |
2 |
2 |
1 |
34 |
166.244 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
44 |
0.38 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.27 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1300 |
0.31 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
9.51 |
-44.89 |
3 |
5 |
1 |
58 |
371.48 |
7 |
↓
|
|
|
|
Analogs
-
38482183
-
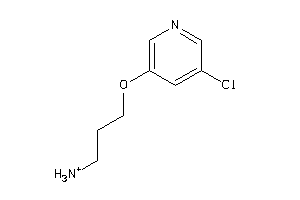
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
140 |
0.87 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.55 |
0.92 |
-46.34 |
3 |
3 |
1 |
50 |
173.623 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
140 |
0.74 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
5.05 |
-36.64 |
1 |
3 |
1 |
27 |
181.259 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.01 |
5.3 |
-83.49 |
2 |
3 |
2 |
28 |
182.267 |
4 |
↓
|
|
|
|
Analogs
-
34380715
-
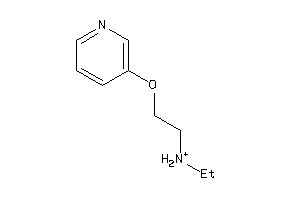
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
35 |
0.95 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
2.31 |
-40.34 |
2 |
3 |
1 |
39 |
153.205 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.71 |
2.59 |
-88.63 |
3 |
3 |
2 |
40 |
154.213 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
4.9 |
-39.55 |
1 |
3 |
1 |
27 |
199.249 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
4.9 |
-39.59 |
1 |
3 |
1 |
27 |
199.249 |
4 |
↓
|
|
|
|
Analogs
-
34439128
-
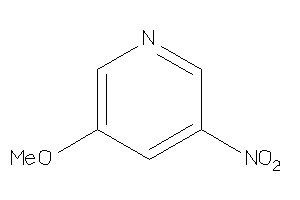
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.81 |
2.95 |
-48.02 |
2 |
6 |
1 |
85 |
198.202 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.81 |
3.24 |
-106.65 |
3 |
6 |
2 |
86 |
199.21 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
50 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
5.05 |
-37.6 |
1 |
3 |
1 |
27 |
246.128 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.91 |
5.34 |
-90.07 |
2 |
3 |
2 |
28 |
247.136 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.83 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHE-1-E |
Acetylcholine Receptor Protein Epsilon Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHG-1-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
Q8BGP7-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.82 |
Functional ≤ 10μM
|
|
ACHA4-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.69 |
Functional ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104281-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
17 |
0.78 |
Binding ≤ 10μM
|
|
Z104284-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta4 (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104285-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104286-2-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104287-3-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #3 Of 3), Other |
Other |
700 |
0.62 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104289-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
8 |
0.81 |
Binding ≤ 10μM
|
|
Z104282-1-O |
Acetylcholine Receptor; Alpha1/beta1/delta/gamma (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 2), Other |
Other |
50 |
0.73 |
Functional ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 2), Other |
Other |
16 |
0.78 |
Functional ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 1), Other |
Other |
28 |
0.76 |
Functional ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 1), Other |
Other |
1 |
0.90 |
Functional ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 1), Other |
Other |
14 |
0.79 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
37 |
0.74 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
7 |
0.82 |
Functional ≤ 10μM
|
|
Z80545-3-O |
TE-671 (cluster #3 Of 3), Other |
Other |
1260 |
0.59 |
Functional ≤ 10μM
|
|
Z81074-1-O |
K177cell Line (cluster #1 Of 1), Other |
Other |
350 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
4.97 |
-42.26 |
2 |
2 |
1 |
29 |
209.7 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHE-1-E |
Acetylcholine Receptor Protein Epsilon Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
ACHG-1-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Binding ≤ 10μM
|
|
Q8BGP7-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.82 |
Functional ≤ 10μM
|
|
ACHA4-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.69 |
Functional ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104281-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
17 |
0.78 |
Binding ≤ 10μM
|
|
Z104284-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta4 (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104285-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104286-2-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104287-3-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #3 Of 3), Other |
Other |
700 |
0.62 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104289-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
8 |
0.81 |
Binding ≤ 10μM
|
|
Z104282-1-O |
Acetylcholine Receptor; Alpha1/beta1/delta/gamma (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 2), Other |
Other |
50 |
0.73 |
Functional ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 2), Other |
Other |
16 |
0.78 |
Functional ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 1), Other |
Other |
28 |
0.76 |
Functional ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 1), Other |
Other |
14 |
0.79 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
37 |
0.74 |
Functional ≤ 10μM
|
|
Z80175-1-O |
IMR-32 (Neuroblastoma Cells) (cluster #1 Of 2), Other |
Other |
7 |
0.82 |
Functional ≤ 10μM
|
|
Z80545-3-O |
TE-671 (cluster #3 Of 3), Other |
Other |
1260 |
0.59 |
Functional ≤ 10μM
|
|
Z81074-1-O |
K177cell Line (cluster #1 Of 1), Other |
Other |
350 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
4.48 |
-44.23 |
2 |
2 |
1 |
29 |
209.7 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4340 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
6.45 |
-9.08 |
0 |
4 |
0 |
44 |
310.397 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.23 |
7.29 |
-34.14 |
1 |
4 |
1 |
45 |
311.405 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4340 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
6.38 |
-7.47 |
0 |
4 |
0 |
44 |
310.397 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.23 |
7.21 |
-32.57 |
1 |
4 |
1 |
45 |
311.405 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
80 |
0.45 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
170 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
8.58 |
-28.64 |
1 |
3 |
1 |
30 |
292.406 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 73 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA2-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3509 |
0.76 |
Binding ≤ 10μM
|
|
ACHB2-3-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHB4-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHD-2-E |
Acetylcholine Receptor Protein Delta Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.74 |
Binding ≤ 10μM
|
|
ACHP-1-E |
Acetylcholine-binding Protein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3162 |
0.77 |
Binding ≤ 10μM
|
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
770 |
0.86 |
Binding ≤ 10μM
|
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.93 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
790 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8900 |
0.71 |
Functional ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.76 |
Functional ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.26 |
Functional ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.93 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
1.19 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
1.12 |
Functional ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
630 |
0.87 |
Functional ≤ 10μM
|
|
Z104281-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #1 Of 2), Other |
Other |
41 |
1.03 |
Binding ≤ 10μM
|
|
Z104286-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #1 Of 2), Other |
Other |
11 |
1.11 |
Binding ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 3), Other |
Other |
881 |
0.85 |
Binding ≤ 10μM
|
|
Z104289-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 2), Other |
Other |
83 |
0.99 |
Binding ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
8 |
1.13 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
12 |
1.11 |
Binding ≤ 10μM
|
|
Z102306-2-O |
Aorta (cluster #2 Of 6), Other |
Other |
1000 |
0.84 |
Functional ≤ 10μM |
|
Z104287-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
8700 |
0.71 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.56 |
6.42 |
-34.1 |
0 |
3 |
1 |
26 |
146.21 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3700 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
2.03 |
-41.43 |
1 |
6 |
1 |
55 |
438.983 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
56446
-
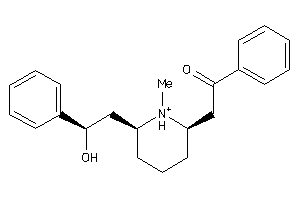
-
156831
-
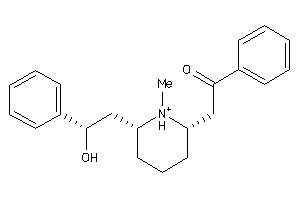
-
156931
-
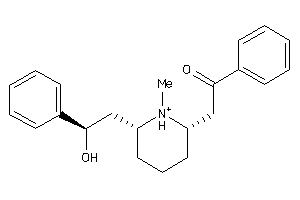
-
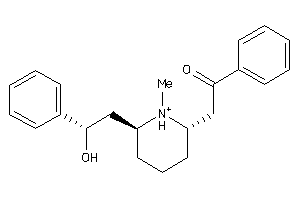
519487
-
-
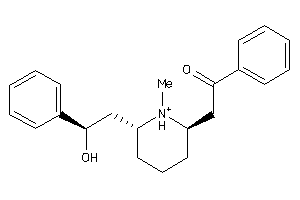
1081316
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
87 |
0.40 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.31 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.47 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
40 |
0.41 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
10.48 |
-45.25 |
2 |
3 |
1 |
42 |
338.471 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.73 |
8.25 |
-8.89 |
1 |
3 |
0 |
41 |
337.463 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA2-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB2-6-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
|
ACHB4-6-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
8.25 |
-39.49 |
3 |
3 |
1 |
45 |
340.487 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
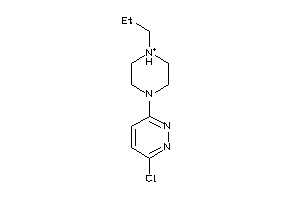
34938206
-
-
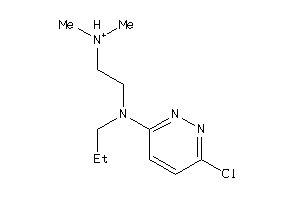
41684221
-
-
42796246
-
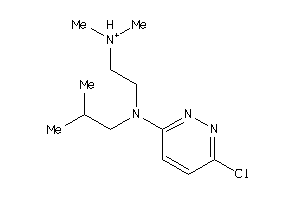
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Binding ≤ 10μM
|
|
ACHB4-6-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
5.36 |
-42.54 |
1 |
4 |
1 |
33 |
227.719 |
1 |
↓
|
|
|
|
Analogs
-
4897762
-
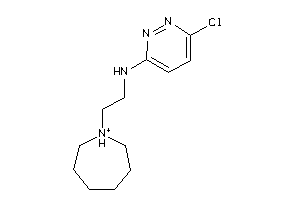
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.78 |
Binding ≤ 10μM
|
|
ACHB4-6-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.78 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4100 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
4.48 |
-48.25 |
2 |
4 |
1 |
46 |
225.703 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
220 |
0.41 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
84 |
0.43 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
10000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
-1.23 |
-8.69 |
0 |
4 |
0 |
43 |
308.381 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
4.33 |
-39.72 |
2 |
3 |
1 |
43 |
167.232 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
322 |
0.57 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
ACHG-1-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3540 |
0.48 |
Binding ≤ 10μM
|
|
Z104282-1-O |
Acetylcholine Receptor; Alpha1/beta1/delta/gamma (cluster #1 Of 1), Other |
Other |
8200 |
0.45 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
617 |
0.54 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
240 |
0.58 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
-2.77 |
-43.65 |
2 |
3 |
1 |
42 |
212.276 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.37 |
14.31 |
-14.39 |
0 |
6 |
0 |
76 |
526.592 |
1 |
↓
|
|
|
|
Analogs
-
5161973
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
3570 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
2.01 |
-31.14 |
2 |
3 |
1 |
39 |
148.189 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.43 |
2.29 |
-90.58 |
3 |
3 |
2 |
40 |
149.197 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
6.06 |
-31.2 |
1 |
3 |
1 |
30 |
202.281 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.44 |
6.34 |
-83.27 |
2 |
3 |
2 |
31 |
203.289 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
47 |
0.93 |
Binding ≤ 10μM
|
|
ACHP-2-E |
Acetylcholine-binding Protein (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3162 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.91 |
1.46 |
-39.03 |
1 |
2 |
1 |
17 |
151.233 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.91 |
1.57 |
-88.04 |
2 |
2 |
2 |
18 |
152.241 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.59 |
4.03 |
-31.32 |
2 |
3 |
1 |
42 |
164.232 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.98 |
4 |
-8.18 |
2 |
3 |
0 |
42 |
163.224 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA2-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA4-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA5-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA6-2-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHA9-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHB-4-E |
Acetylcholine Receptor Protein Beta Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHB2-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHB3-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
|
ACHB4-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
1400 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
1.65 |
-32.73 |
2 |
4 |
1 |
48 |
178.215 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.49 |
1.92 |
-85.07 |
3 |
4 |
2 |
49 |
179.223 |
3 |
↓
|
|
|
|
Analogs
-
53132543
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
1510 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.03 |
1.29 |
-34.74 |
4 |
3 |
1 |
65 |
136.178 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.03 |
1.58 |
-91.14 |
5 |
3 |
2 |
66 |
137.186 |
2 |
↓
|
|
|
|
Analogs
-
519487
-

-
1624
-
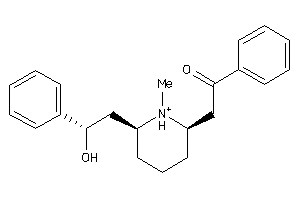
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
6260 |
0.29 |
Binding ≤ 10μM
|
|
VMAT2-1-E |
Synaptic Vesicular Amine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.31 |
Binding ≤ 10μM
|
|
CP2D6-3-E |
Cytochrome P450 2D6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
10.67 |
-44.31 |
2 |
3 |
1 |
42 |
338.471 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.73 |
8.5 |
-7.31 |
1 |
3 |
0 |
41 |
337.463 |
6 |
↓
|
|
|
|
Analogs
-
4245635
-
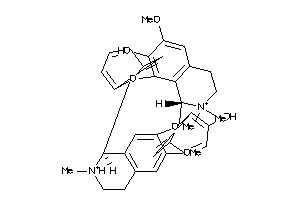
-
38153422
-
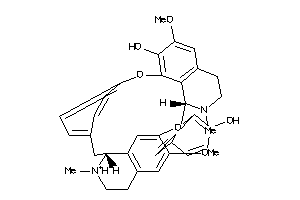
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4790 |
0.17 |
Binding ≤ 10μM
|
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.17 |
Binding ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.17 |
Binding ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.17 |
Binding ≤ 10μM
|
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.16 |
Functional ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.16 |
Functional ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.16 |
Functional ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
9720 |
0.16 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
2000 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
3.03 |
-94.42 |
3 |
8 |
2 |
81 |
610.751 |
2 |
↓
|
|
|
|
Analogs
-
3704277
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
6.34 |
-35.48 |
1 |
2 |
1 |
14 |
192.282 |
2 |
↓
|
|
|
|
Analogs
-
3704277
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA-2-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
707 |
0.62 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
6.94 |
-35.04 |
1 |
2 |
1 |
14 |
192.282 |
2 |
↓
|
|
|
|
Analogs
-
1912635
-
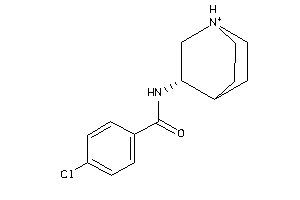
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1662 |
0.45 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1662 |
0.45 |
Binding ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1662 |
0.45 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1662 |
0.45 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1662 |
0.45 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
27 |
0.59 |
Binding ≤ 10μM
|
|
ACHP-3-E |
Acetylcholine-binding Protein (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2089 |
0.44 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
154 |
0.53 |
Functional ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4541 |
0.42 |
Functional ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
154 |
0.53 |
Functional ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
154 |
0.53 |
Functional ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
154 |
0.53 |
Functional ≤ 10μM
|
|
ACHA7-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
154 |
0.53 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
4541 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
6.99 |
-40.22 |
2 |
3 |
1 |
34 |
265.764 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
120 |
0.54 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1950 |
0.44 |
Binding ≤ 10μM
|
|
ACHP-2-E |
Acetylcholine-binding Protein (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7943 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
-0.87 |
-37.46 |
2 |
4 |
1 |
43 |
247.318 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
120 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
-0.88 |
-37.56 |
2 |
4 |
1 |
43 |
247.318 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
33 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
9.86 |
-44.69 |
2 |
5 |
1 |
60 |
362.453 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.90 |
10.32 |
-107.06 |
3 |
5 |
2 |
61 |
363.461 |
4 |
↓
|
|
|
|
Analogs
-
34463140
-
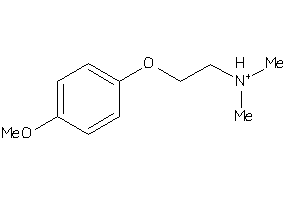
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1100 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.95 |
3.81 |
-31.73 |
0 |
2 |
1 |
9 |
180.271 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
511 |
0.44 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
511 |
0.44 |
Binding ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
511 |
0.44 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
511 |
0.44 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
511 |
0.44 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.56 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.50 |
Functional ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.50 |
Functional ≤ 10μM
|
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.50 |
Functional ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.50 |
Functional ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.50 |
Functional ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
65 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
5.15 |
-40.57 |
2 |
5 |
1 |
60 |
272.328 |
2 |
↓
|
|
|
|
Analogs
-
4098884
-
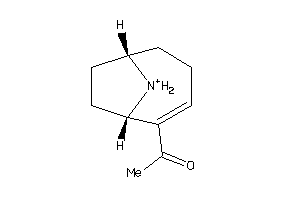
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB-3-E |
Acetylcholine Receptor Protein Beta Chain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.94 |
Binding ≤ 10μM
|
|
ACHG-3-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.81 |
Functional ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
48 |
0.85 |
Binding ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2000 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
-0.69 |
-42.22 |
2 |
2 |
1 |
33 |
166.244 |
1 |
↓
|
|
|
|
Analogs
-
391820
-
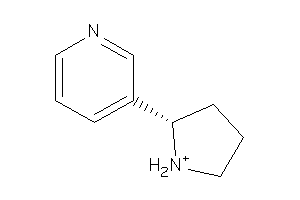
Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.83 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
1.08 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
730 |
0.78 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.83 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
17 |
0.99 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.90 |
Binding ≤ 10μM
|
|
ACHG-1-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.83 |
Binding ≤ 10μM
|
|
Q8BGP7-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
1.08 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
29 |
0.96 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
30 |
0.96 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
3.72 |
-39.03 |
2 |
2 |
1 |
29 |
149.217 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.85 |
4 |
-97.03 |
3 |
2 |
2 |
31 |
150.225 |
1 |
↓
|
|
|
|
Analogs
-
3786099
-
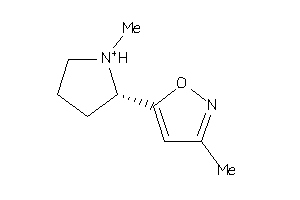
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM |
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM |
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.98 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
5.81 |
-35.61 |
1 |
3 |
1 |
30 |
167.232 |
1 |
↓
|
|
|
|
Analogs
-
8089
-
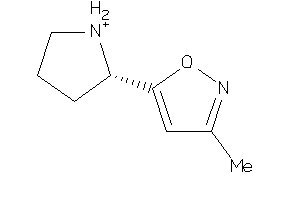
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
3.56 |
-40.1 |
2 |
3 |
1 |
43 |
153.205 |
1 |
↓
|
|