|
|
Analogs
-
4051004
-
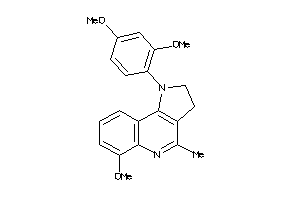
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.34 |
Binding ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
230 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
8.96 |
-22.49 |
1 |
4 |
1 |
36 |
321.4 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 79 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
4.06 |
-43.78 |
2 |
6 |
1 |
78 |
346.432 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.41 |
3.62 |
-13.05 |
1 |
6 |
0 |
77 |
345.424 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 87 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
3.99 |
-43.94 |
2 |
6 |
1 |
78 |
346.432 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.41 |
3.56 |
-24.45 |
1 |
6 |
0 |
77 |
345.424 |
5 |
↓
|
|
|
|
Analogs
-
4051001
-
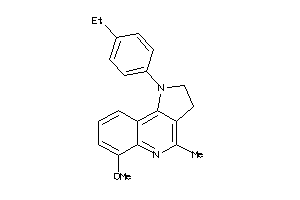
-
4050991
-
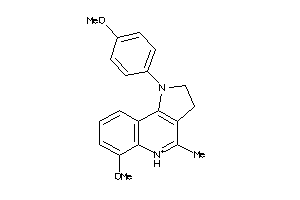
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.36 |
Binding ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
84 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
9.55 |
-22.04 |
1 |
3 |
1 |
27 |
291.374 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1320 |
0.43 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1320 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
9.04 |
-9.08 |
3 |
4 |
0 |
64 |
250.305 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.34 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.37 |
0.68 |
-9.93 |
2 |
4 |
0 |
49 |
326.403 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.48 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.48 |
Binding ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.34 |
ADME/T ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
13 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
9.9 |
-17.91 |
0 |
4 |
0 |
50 |
277.327 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.33 |
10.35 |
-35.25 |
1 |
4 |
1 |
52 |
278.335 |
4 |
↓
|
|
|
|
Analogs
-
4192432
-
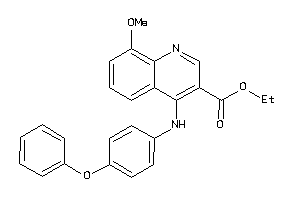
-
4192441
-
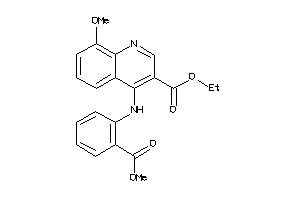
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3540 |
0.32 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3540 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
8.65 |
-9.96 |
1 |
5 |
0 |
60 |
322.364 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.36 |
9.14 |
-31.09 |
2 |
5 |
1 |
62 |
323.372 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1430 |
0.45 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1430 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
8.01 |
-5.92 |
1 |
2 |
0 |
25 |
234.302 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.40 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
8.6 |
-9.06 |
2 |
4 |
0 |
55 |
250.305 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.34 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.77 |
1.29 |
-9.81 |
2 |
4 |
0 |
49 |
340.43 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1390 |
0.39 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1390 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
11 |
-9.09 |
2 |
4 |
0 |
50 |
278.359 |
4 |
↓
|
|