|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
7 |
0.42 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1500 |
0.30 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.45 |
Binding ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
27 |
0.39 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
45 |
0.38 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.40 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
770 |
0.32 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
2 |
0.45 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
3 |
0.44 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
7 |
0.42 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
6 |
0.43 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
11 |
0.41 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
16 |
0.40 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
5 |
0.43 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.37 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
58 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
5.85 |
-37.21 |
6 |
8 |
1 |
119 |
370.433 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.19 |
5.38 |
-15.09 |
5 |
8 |
0 |
118 |
369.425 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-3-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
DYR-2-E |
Dihydrofolate Reductase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
-4.73 |
-7.48 |
4 |
7 |
0 |
98 |
394.69 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-3-E |
Dihydrofolate Reductase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2 |
0.47 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
3 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
-2.68 |
-13.82 |
4 |
7 |
0 |
105 |
354.41 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.39 |
-2.49 |
-43.52 |
5 |
7 |
1 |
106 |
355.418 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-3-E |
Dihydrofolate Reductase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
3 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
-2.65 |
-13.66 |
4 |
7 |
0 |
105 |
354.41 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.39 |
-2.46 |
-41.86 |
5 |
7 |
1 |
106 |
355.418 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.37 |
4.29 |
-128.65 |
5 |
13 |
-2 |
219 |
439.388 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.91 |
2.11 |
-174.25 |
4 |
13 |
-3 |
222 |
438.38 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.37 |
2.32 |
-71.85 |
6 |
13 |
-1 |
216 |
440.396 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2370 |
0.23 |
Binding ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
740 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
4.76 |
-126.34 |
6 |
14 |
-2 |
225 |
471.43 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
-3.04 |
4.11 |
-136.66 |
6 |
14 |
-2 |
225 |
471.43 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.04 |
2.09 |
-83.69 |
7 |
14 |
-1 |
223 |
472.438 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2370 |
0.23 |
Binding ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
740 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
4.86 |
-126.36 |
6 |
14 |
-2 |
225 |
471.43 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.04 |
2.16 |
-83.91 |
7 |
14 |
-1 |
223 |
472.438 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.04 |
2.88 |
-72.52 |
7 |
14 |
-1 |
223 |
472.438 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
52 |
0.24 |
Binding ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
650 |
0.21 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
10 |
0.27 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80192-1-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
3 |
0.28 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4 |
0.28 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.28 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
5 |
0.28 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
46 |
0.24 |
Functional ≤ 10μM
|
|
Z80356-1-O |
OVCAR-5 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.28 |
Functional ≤ 10μM
|
|
Z80357-1-O |
OVCAR-8 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.29 |
Functional ≤ 10μM
|
|
Z80438-1-O |
RXF 393 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
9 |
0.27 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
5 |
0.28 |
Functional ≤ 10μM
|
|
Z80488-2-O |
SK-MEL-5 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
6 |
0.27 |
Functional ≤ 10μM
|
|
Z80551-1-O |
TK-10 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.27 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
5 |
0.28 |
Functional ≤ 10μM
|
|
Z80568-1-O |
UACC-257 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
25 |
0.25 |
Functional ≤ 10μM
|
|
Z80570-3-O |
UACC-62 (Melanoma Cells) (cluster #3 Of 3), Other |
Other |
2 |
0.29 |
Functional ≤ 10μM
|
|
Z80575-1-O |
UO-31 (Renal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
6 |
0.27 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.28 |
Functional ≤ 10μM
|
|
Z80834-1-O |
EKVX (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.25 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
440 |
0.21 |
Functional ≤ 10μM
|
|
Z80920-1-O |
HCC 2998 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80972-1-O |
HOP-92 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
8 |
0.27 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
3 |
0.28 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
5 |
0.28 |
Functional ≤ 10μM |
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.28 |
Functional ≤ 10μM
|
|
Z81248-1-O |
Malme-3M (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.29 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
4 |
0.28 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
7 |
0.27 |
Functional ≤ 10μM
|
|
Z81322-1-O |
SF-539 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
4 |
0.28 |
Functional ≤ 10μM
|
|
Z81325-1-O |
SR (Leukemia Cells) (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
2 |
0.29 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
1 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.11 |
-10.62 |
-142.86 |
7 |
15 |
-2 |
254 |
571.554 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
4.57 |
-8.9 |
4 |
5 |
0 |
87 |
230.271 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.40 |
1.17 |
-31.96 |
5 |
5 |
1 |
88 |
231.279 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.40 |
4.87 |
-32.69 |
5 |
5 |
1 |
88 |
231.279 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.34 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3620 |
0.36 |
Binding ≤ 10μM
|
|
B0BL08-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
10 |
0.53 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2700 |
0.37 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
20 |
0.51 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
300 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
152 |
0.45 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
103 |
0.47 |
Functional ≤ 10μM
|
|
Z50424-1-O |
Cryptosporidium Parvum (cluster #1 Of 2), Other |
Other |
4000 |
0.36 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
9309 |
0.34 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
4.4 |
-13.71 |
4 |
7 |
0 |
106 |
290.323 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.00 |
4.89 |
-42.71 |
5 |
7 |
1 |
107 |
291.331 |
5 |
↓
|
|
|
|
Analogs
-
1851132
-
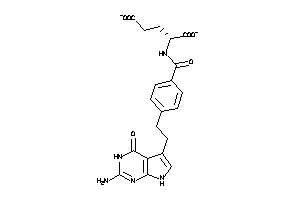
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.29 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.29 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2300 |
0.25 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1100 |
0.27 |
Binding ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
16 |
0.35 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
22 |
0.35 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
894 |
0.27 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
68 |
0.32 |
Functional ≤ 10μM |
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
894 |
0.27 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.53 |
8.7 |
-125.85 |
5 |
11 |
-2 |
197 |
425.401 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.53 |
6.72 |
-70.62 |
6 |
11 |
-1 |
194 |
426.409 |
9 |
↓
|
|
|
|
Analogs
-
4243472
-
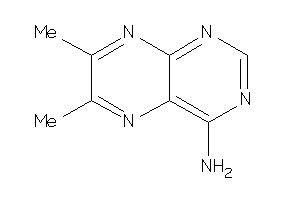
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9100 |
0.50 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
3900 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.02 |
1.36 |
-12.22 |
4 |
6 |
0 |
104 |
190.21 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.02 |
1.67 |
-25.18 |
5 |
6 |
1 |
105 |
191.218 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.02 |
-2.42 |
-30.82 |
5 |
6 |
1 |
105 |
191.218 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
690 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
330 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
3.87 |
-6 |
4 |
7 |
0 |
128 |
289.73 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.81 |
4.16 |
-30.86 |
5 |
7 |
1 |
129 |
290.738 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.81 |
0.52 |
-28.67 |
5 |
7 |
1 |
129 |
290.738 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.17 |
-6.48 |
-141.4 |
5 |
12 |
-2 |
212 |
449.427 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.17 |
-6.28 |
-157.02 |
6 |
12 |
-1 |
214 |
450.435 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.17 |
-6.26 |
-146.89 |
6 |
12 |
-1 |
214 |
450.435 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3,5-diamino-7-methyl-2,4,10-triazabicyclo[4.4.0]deca-2,4,6,8,10-pentaen-8-yl)methanol
(3,5-diamino-7-methyl-2,4,10-tri…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
0.24 |
-15.83 |
5 |
6 |
0 |
111 |
205.221 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.26 |
-5.17 |
-33.44 |
6 |
6 |
1 |
112 |
206.229 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.26 |
0.57 |
-28.19 |
6 |
6 |
1 |
112 |
206.229 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-3-E |
Dihydrofolate Reductase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
-3.54 |
-12.4 |
4 |
6 |
0 |
96 |
339.193 |
4 |
↓
|
|
|
|
Analogs
-
6920406
-
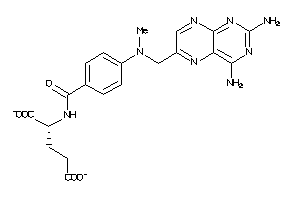
Draw
Identity
99%
90%
80%
70%
Vendors
And 72 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B0BL08-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
7 |
0.35 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
7 |
0.35 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
Q81R22-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
15 |
0.33 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1800 |
0.24 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
26 |
0.32 |
Functional ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.34 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
33 |
0.32 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.35 |
Binding ≤ 10μM
|
|
FOLC-1-E |
Folylpoly-gamma-glutamate Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
39 |
0.31 |
Binding ≤ 10μM
|
|
S19A1-1-E |
Folate Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.23 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3600 |
0.23 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
880 |
0.26 |
Binding ≤ 10μM
|
|
Z103204-2-O |
A427 (cluster #2 Of 4), Other |
Other |
5520 |
0.22 |
Functional ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
90 |
0.30 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
85 |
0.30 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6600 |
0.22 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4200 |
0.23 |
Functional ≤ 10μM
|
|
Z80001-1-O |
143B (cluster #1 Of 2), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.31 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
1100 |
0.25 |
Functional ≤ 10μM
|
|
Z80063-1-O |
CCRF S-180 (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
620 |
0.26 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
870 |
0.26 |
Functional ≤ 10μM
|
|
Z80112-1-O |
DAN-G (cluster #1 Of 2), Other |
Other |
77 |
0.30 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
45 |
0.31 |
Functional ≤ 10μM
|
|
Z80132-1-O |
EL4 (Thymoma Cells) (cluster #1 Of 2), Other |
Other |
1570 |
0.25 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.35 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80166-12-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #12 Of 12), Other |
Other |
4400 |
0.23 |
Functional ≤ 10μM |
|
Z80171-1-O |
HuTu80 (cluster #1 Of 1), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z80192-1-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
42 |
0.31 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80214-1-O |
M14 (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
32 |
0.32 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
900 |
0.26 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
400 |
0.27 |
Functional ≤ 10μM
|
|
Z80356-1-O |
OVCAR-5 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
980 |
0.25 |
Functional ≤ 10μM
|
|
Z80357-1-O |
OVCAR-8 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80362-8-O |
P388 (Lymphoma Cells) (cluster #8 Of 8), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM |
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
27 |
0.32 |
Functional ≤ 10μM
|
|
Z80407-1-O |
R1 (cluster #1 Of 1), Other |
Other |
720 |
0.26 |
Functional ≤ 10μM
|
|
Z80408-1-O |
R1-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
985 |
0.25 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
216 |
0.28 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
2600 |
0.24 |
Functional ≤ 10μM
|
|
Z80410-1-O |
R2-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
2630 |
0.24 |
Functional ≤ 10μM
|
|
Z80411-1-O |
R30dm-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
18 |
0.33 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.31 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM
|
|
Z80455-1-O |
SCC-15 (cluster #1 Of 1), Other |
Other |
580 |
0.26 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
36 |
0.32 |
Functional ≤ 10μM
|
|
Z80488-2-O |
SK-MEL-5 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
87 |
0.30 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
63 |
0.31 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
12 |
0.34 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
22 |
0.32 |
Functional ≤ 10μM
|
|
Z80568-1-O |
UACC-257 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
790 |
0.26 |
Functional ≤ 10μM
|
|
Z80570-3-O |
UACC-62 (Melanoma Cells) (cluster #3 Of 3), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80575-1-O |
UO-31 (Renal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
191 |
0.29 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80657-1-O |
A253 Cell Line (cluster #1 Of 1), Other |
Other |
3000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM |
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
37 |
0.32 |
Functional ≤ 10μM |
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80752-1-O |
Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80789-1-O |
Colon 38 (cluster #1 Of 1), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
13 |
0.33 |
Functional ≤ 10μM
|
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
9740 |
0.21 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
906 |
0.26 |
Functional ≤ 10μM
|
|
Z80893-1-O |
H35 (Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80920-1-O |
HCC 2998 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
110 |
0.30 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
25 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
52 |
0.31 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
65 |
0.30 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM |
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
100 |
0.30 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
43 |
0.31 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
450 |
0.27 |
Functional ≤ 10μM
|
|
Z81322-1-O |
SF-539 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.32 |
Functional ≤ 10μM
|
|
Z81325-1-O |
SR (Leukemia Cells) (cluster #1 Of 2), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z80253-1-O |
MEF (Embryonic Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
120 |
0.29 |
ADME/T ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
216 |
0.28 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.97 |
6.06 |
-125.84 |
5 |
13 |
-2 |
216 |
452.431 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.97 |
6.52 |
-124.82 |
6 |
13 |
-1 |
217 |
453.439 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
5.5 |
-8.97 |
4 |
5 |
0 |
87 |
244.298 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.80 |
5.8 |
-34.52 |
5 |
5 |
1 |
88 |
245.306 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.80 |
2.09 |
-33.23 |
5 |
5 |
1 |
88 |
245.306 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.25 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2600 |
0.22 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5520 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.19 |
6.58 |
-135.09 |
4 |
12 |
-2 |
208 |
476.449 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.74 |
5.48 |
-177.48 |
3 |
12 |
-3 |
211 |
475.441 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.19 |
5.46 |
-83.12 |
5 |
12 |
-1 |
205 |
477.457 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.56 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
23 |
0.71 |
Binding ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
1450 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
3.47 |
-26.38 |
6 |
5 |
1 |
95 |
200.225 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.12 |
2.99 |
-11.68 |
5 |
5 |
0 |
94 |
199.217 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.46 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Dihydrofolate Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Functional ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
10 |
0.66 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
80 |
0.58 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
9900 |
0.41 |
Functional ≤ 10μM |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
700 |
0.51 |
Functional ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
5000 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
6.53 |
-28.58 |
5 |
4 |
1 |
79 |
249.725 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.84 |
6.08 |
-5.33 |
4 |
4 |
0 |
78 |
248.717 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
35 |
0.33 |
Binding ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
320 |
0.28 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
|
Z80119-1-O |
Detroit 98 (cluster #1 Of 1), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
|
Z80171-1-O |
HuTu80 (cluster #1 Of 1), Other |
Other |
5 |
0.36 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7900 |
0.22 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
7 |
0.36 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2050 |
0.25 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
320 |
0.28 |
Functional ≤ 10μM
|
|
Z81131-1-O |
L Cells (Fibroblasts) (cluster #1 Of 1), Other |
Other |
2 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
4.48 |
-122.21 |
6 |
13 |
-2 |
225 |
438.404 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.21 |
4.93 |
-127.62 |
7 |
13 |
-1 |
226 |
439.412 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C3SWJ7-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
19 |
0.31 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
22 |
0.31 |
Binding ≤ 10μM
|
|
E2QDN0-1-B |
Thymidylate Synthase (EC 2.1.1.45) (TS) (TSase) (cluster #1 Of 1), Bacterial |
Bacteria |
48 |
0.29 |
Binding ≤ 10μM |
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
90 |
0.28 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
95 |
0.28 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.29 |
Binding ≤ 10μM |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.25 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.21 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
Z100337-1-O |
W1-L2 (cluster #1 Of 1), Other |
Other |
440 |
0.25 |
Functional ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
750 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5 |
0.33 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
60 |
0.29 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7000 |
0.21 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.23 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.23 |
Functional ≤ 10μM
|
|
Z80860-1-O |
GC3/M Cell Line (cluster #1 Of 1), Other |
Other |
2130 |
0.23 |
Functional ≤ 10μM
|
|
Z80862-1-O |
GC3/MTK- Cell Line (cluster #1 Of 1), Other |
Other |
1400 |
0.23 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z81133-1-O |
L1210 (1565) (cluster #1 Of 1), Other |
Other |
5500 |
0.21 |
Functional ≤ 10μM
|
|
Z81221-1-O |
Lymphoma Cell Line (cluster #1 Of 1), Other |
Other |
560 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
10.16 |
-129.42 |
4 |
11 |
-2 |
184 |
475.461 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.36 |
8.12 |
-191.26 |
3 |
11 |
-3 |
187 |
474.453 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.82 |
8.22 |
-71.83 |
5 |
11 |
-1 |
182 |
476.469 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
22 |
0.31 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
90 |
0.28 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
95 |
0.28 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
430 |
0.25 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PUR2-1-E |
GAR Transformylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.21 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.26 |
Binding ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
750 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
5 |
0.33 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7000 |
0.21 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
1730 |
0.23 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1820 |
0.23 |
Functional ≤ 10μM
|
|
Z80860-1-O |
GC3/M Cell Line (cluster #1 Of 1), Other |
Other |
2130 |
0.23 |
Functional ≤ 10μM
|
|
Z80862-1-O |
GC3/MTK- Cell Line (cluster #1 Of 1), Other |
Other |
1400 |
0.23 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z81221-1-O |
Lymphoma Cell Line (cluster #1 Of 1), Other |
Other |
560 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
8.37 |
-132.31 |
4 |
11 |
-2 |
184 |
475.461 |
10 |
↓
|
|
|
|
Analogs
-
1529323
-
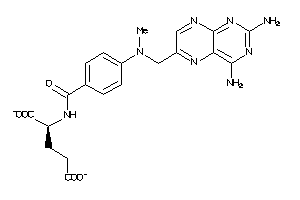
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B0BL08-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
9 |
0.34 |
Binding ≤ 10μM
|
|
Q81R22-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
15 |
0.33 |
Binding ≤ 10μM
|
|
TYSY-1-B |
Thymidylate Synthase (cluster #1 Of 1), Bacterial |
Bacteria |
1800 |
0.24 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
26 |
0.32 |
Functional ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
33 |
0.32 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.35 |
Binding ≤ 10μM
|
|
FOLC-1-E |
Folylpoly-gamma-glutamate Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
PTR1-1-E |
Pteridine Reductase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
39 |
0.31 |
Binding ≤ 10μM
|
|
S19A1-1-E |
Folate Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.23 |
Binding ≤ 10μM
|
|
TYSY-1-E |
Thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3600 |
0.23 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
880 |
0.26 |
Binding ≤ 10μM
|
|
Z103204-2-O |
A427 (cluster #2 Of 4), Other |
Other |
5520 |
0.22 |
Functional ≤ 10μM
|
|
Z50266-1-O |
Enterococcus Faecium (cluster #1 Of 1), Other |
Other |
90 |
0.30 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
85 |
0.30 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6600 |
0.22 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4200 |
0.23 |
Functional ≤ 10μM
|
|
Z80001-1-O |
143B (cluster #1 Of 2), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
40 |
0.31 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
1100 |
0.25 |
Functional ≤ 10μM
|
|
Z80063-1-O |
CCRF S-180 (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
870 |
0.26 |
Functional ≤ 10μM
|
|
Z80112-1-O |
DAN-G (cluster #1 Of 2), Other |
Other |
77 |
0.30 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
45 |
0.31 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80166-12-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #12 Of 12), Other |
Other |
4400 |
0.23 |
Functional ≤ 10μM |
|
Z80171-1-O |
HuTu80 (cluster #1 Of 1), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z80192-1-O |
KM12 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
42 |
0.31 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80214-1-O |
M14 (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
32 |
0.32 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
900 |
0.26 |
Functional ≤ 10μM
|
|
Z80227-1-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
400 |
0.27 |
Functional ≤ 10μM
|
|
Z80356-1-O |
OVCAR-5 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
980 |
0.25 |
Functional ≤ 10μM
|
|
Z80357-1-O |
OVCAR-8 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80362-8-O |
P388 (Lymphoma Cells) (cluster #8 Of 8), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM |
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
27 |
0.32 |
Functional ≤ 10μM
|
|
Z80407-1-O |
R1 (cluster #1 Of 1), Other |
Other |
720 |
0.26 |
Functional ≤ 10μM
|
|
Z80408-1-O |
R1-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
985 |
0.25 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
2600 |
0.24 |
Functional ≤ 10μM
|
|
Z80410-1-O |
R2-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
2630 |
0.24 |
Functional ≤ 10μM
|
|
Z80411-1-O |
R30dm-CCRF-CEM (Lymphoblastic Leukemia) (cluster #1 Of 1), Other |
Other |
18 |
0.33 |
Functional ≤ 10μM
|
|
Z80433-1-O |
RPMI-8226 (Multiple Myeloma Cells) (cluster #1 Of 3), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80443-1-O |
S180 (Sarcoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM
|
|
Z80455-1-O |
SCC-15 (cluster #1 Of 1), Other |
Other |
580 |
0.26 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM
|
|
Z80468-1-O |
SF-295 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
36 |
0.32 |
Functional ≤ 10μM
|
|
Z80488-2-O |
SK-MEL-5 (Melanoma Cells) (cluster #2 Of 3), Other |
Other |
87 |
0.30 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80560-1-O |
U-251 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
63 |
0.31 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
12 |
0.34 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
22 |
0.32 |
Functional ≤ 10μM
|
|
Z80568-1-O |
UACC-257 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
790 |
0.26 |
Functional ≤ 10μM
|
|
Z80570-3-O |
UACC-62 (Melanoma Cells) (cluster #3 Of 3), Other |
Other |
28 |
0.32 |
Functional ≤ 10μM
|
|
Z80575-1-O |
UO-31 (Renal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
191 |
0.29 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
9 |
0.34 |
Functional ≤ 10μM
|
|
Z80640-1-O |
786-0 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z80657-1-O |
A253 Cell Line (cluster #1 Of 1), Other |
Other |
3000 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
5 |
0.35 |
Functional ≤ 10μM |
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
78 |
0.30 |
Functional ≤ 10μM
|
|
Z80752-1-O |
Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
15 |
0.33 |
Functional ≤ 10μM
|
|
Z80789-1-O |
Colon 38 (cluster #1 Of 1), Other |
Other |
66 |
0.30 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
16 |
0.33 |
Functional ≤ 10μM
|
|
Z80830-1-O |
Ehrlich (Ehrlich Ascites Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
13 |
0.33 |
Functional ≤ 10μM
|
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
31 |
0.32 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
9740 |
0.21 |
Functional ≤ 10μM
|
|
Z80890-1-O |
H350.3 (MTX-resistant Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
906 |
0.26 |
Functional ≤ 10μM
|
|
Z80893-1-O |
H35 (Reuber Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z80920-1-O |
HCC 2998 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
110 |
0.30 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
25 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
10 |
0.34 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
52 |
0.31 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
65 |
0.30 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
6 |
0.35 |
Functional ≤ 10μM |
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
100 |
0.30 |
Functional ≤ 10μM
|
|
Z81281-1-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 5), Other |
Other |
43 |
0.31 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
450 |
0.27 |
Functional ≤ 10μM
|
|
Z81322-1-O |
SF-539 (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
35 |
0.32 |
Functional ≤ 10μM
|
|
Z81325-1-O |
SR (Leukemia Cells) (cluster #1 Of 2), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
33 |
0.32 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
30 |
0.32 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
216 |
0.28 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.97 |
6.03 |
-126.53 |
5 |
13 |
-2 |
216 |
452.431 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.97 |
6.57 |
-124.3 |
6 |
13 |
-1 |
217 |
453.439 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLAT-2-E |
Choline Acetylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.21 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.19 |
Binding ≤ 10μM
|
|
ENPP2-1-E |
Autotaxin (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.17 |
Binding ≤ 10μM
|
|
AMPC-6-B |
Beta-lactamase (cluster #6 Of 6), Bacterial |
Bacteria |
650 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
5.66 |
-122.53 |
4 |
12 |
-2 |
216 |
650.698 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
4517559
-
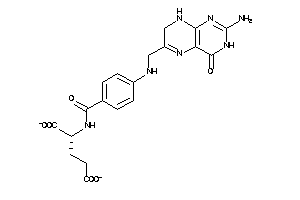
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.50 |
4.26 |
-131.57 |
6 |
13 |
-2 |
218 |
441.404 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
310 |
0.41 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.33 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2800 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
-2.95 |
-8.5 |
4 |
7 |
0 |
105 |
319.8 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CTRC-2-E |
Chymotrypsin C (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
DYR-3-E |
Dihydrofolate Reductase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
AMPC-2-B |
Beta-lactamase (cluster #2 Of 6), Bacterial |
Bacteria |
500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.46 |
-2.82 |
-17.64 |
1 |
5 |
0 |
67 |
473.819 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
91 |
0.35 |
Binding ≤ 10μM
|
|
DYR-2-E |
Dihydrofolate Reductase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
1600 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
12.08 |
-35.97 |
5 |
8 |
1 |
128 |
379.444 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.55 |
11.62 |
-9.15 |
4 |
8 |
0 |
127 |
378.436 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.56 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.55 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
190 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.43 |
4.37 |
-32.03 |
4 |
6 |
1 |
106 |
275.723 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.43 |
1.01 |
-82.26 |
5 |
6 |
2 |
107 |
276.731 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.43 |
4.67 |
-78.48 |
5 |
6 |
2 |
107 |
276.731 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-3-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
82 |
0.58 |
Binding ≤ 10μM
|
|
DYR-2-E |
Dihydrofolate Reductase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
110 |
0.57 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Dihydrofolate Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.43 |
Functional ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
450 |
0.52 |
Binding ≤ 10μM
|
|
Z50425-7-O |
Plasmodium Falciparum (cluster #7 Of 22), Other |
Other |
8 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.90 |
7.06 |
-25.38 |
5 |
5 |
1 |
82 |
252.729 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.16 |
6.71 |
-4.72 |
4 |
5 |
0 |
75 |
251.721 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
38 |
0.43 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.51 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.45 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
30 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
260 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-3.63 |
-16.02 |
4 |
7 |
0 |
109 |
325.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.41 |
-41.04 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.4 |
-28.87 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
940 |
0.42 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
23 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
-3.5 |
-7.62 |
4 |
4 |
0 |
77 |
282.372 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
660 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
8.15 |
-29.58 |
5 |
5 |
1 |
88 |
335.309 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.45 |
7.67 |
-10.58 |
4 |
5 |
0 |
87 |
334.301 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.34 |
-6.43 |
-124.44 |
5 |
12 |
-2 |
212 |
465.47 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.34 |
6.37 |
-122.1 |
5 |
12 |
-2 |
213 |
465.47 |
10 |
↓
|
|