|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1500 |
0.23 |
Binding ≤ 10μM
|
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
479 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.68 |
1.1 |
-19.39 |
1 |
5 |
0 |
72 |
501.733 |
10 |
↓
|
|
|
|
Analogs
-
42077255
-
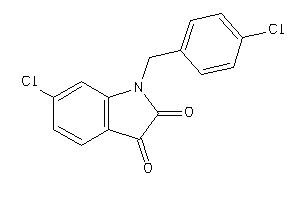
-
42101101
-
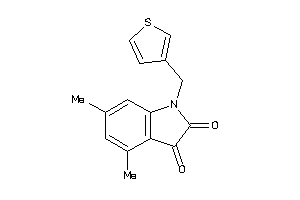
-
42101255
-
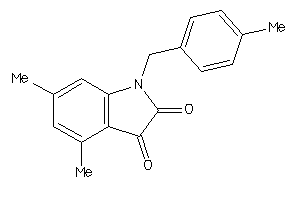
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1700 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
1.38 |
-12.11 |
0 |
3 |
0 |
39 |
237.258 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
23 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
7.45 |
-10.93 |
0 |
3 |
0 |
39 |
223.231 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
FAAH1-5-E |
Anandamide Amidohydrolase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.20 |
14.1 |
-13.02 |
0 |
4 |
0 |
56 |
382.548 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
FAAH1-5-E |
Anandamide Amidohydrolase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
8.61 |
-12.58 |
0 |
4 |
0 |
56 |
294.354 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.19 |
8.87 |
-41.41 |
1 |
4 |
1 |
57 |
295.362 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
600 |
0.35 |
Binding ≤ 10μM
|
|
FAAH1-5-E |
Anandamide Amidohydrolase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
0.21 |
-9.29 |
0 |
4 |
0 |
55 |
334.419 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-2-E |
Carboxylesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3080 |
0.43 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
2.34 |
-6.19 |
0 |
2 |
0 |
34 |
238.286 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
107 |
0.49 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
334 |
0.45 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
10.47 |
-4.87 |
0 |
2 |
0 |
34 |
396.078 |
5 |
↓
|
|
|
|
Analogs
-
4265685
-
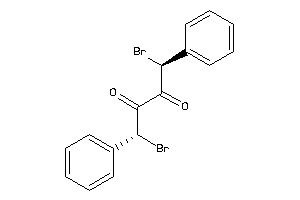
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
107 |
0.49 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
334 |
0.45 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
0.83 |
-4.26 |
0 |
2 |
0 |
34 |
396.078 |
5 |
↓
|
|
|
|
Analogs
-
4265684
-
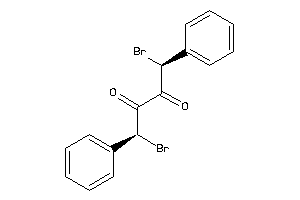
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
107 |
0.49 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
334 |
0.45 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
0.87 |
-4.37 |
0 |
2 |
0 |
34 |
396.078 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
2630 |
0.41 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
267 |
0.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
1.63 |
-5.03 |
0 |
2 |
0 |
34 |
317.182 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
2630 |
0.41 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
267 |
0.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
1.58 |
-5.2 |
0 |
2 |
0 |
34 |
317.182 |
5 |
↓
|
|
|
|
Analogs
-
5819582
-
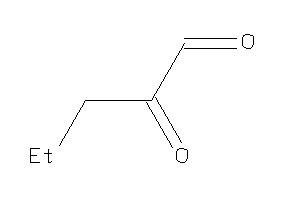
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-2-E |
Carboxylesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8060 |
0.71 |
Binding ≤ 10μM |
|
EST2-2-E |
Carboxylesterase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4670 |
0.75 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
5.02 |
-2.74 |
0 |
2 |
0 |
34 |
142.198 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
1550 |
0.68 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2420 |
0.66 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
5.72 |
-4.15 |
0 |
2 |
0 |
34 |
162.188 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-2-E |
Carboxylesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
98 |
0.65 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
8.05 |
-3.92 |
0 |
2 |
0 |
34 |
204.269 |
6 |
↓
|
|
|
|
Analogs
-
3844983
-
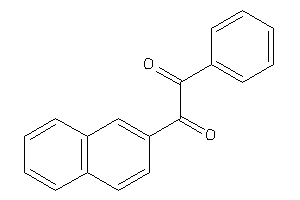
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
257 |
0.38 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
85 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
2.17 |
-8.77 |
0 |
2 |
0 |
34 |
310.352 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
830 |
0.53 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
45 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
1.93 |
-7.72 |
0 |
2 |
0 |
34 |
210.232 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
4540 |
0.53 |
Binding ≤ 10μM
|
|
EST2-5-E |
Carboxylesterase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5610 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
5.48 |
-57.61 |
0 |
2 |
-1 |
40 |
223.298 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
4540 |
0.53 |
Binding ≤ 10μM
|
|
EST2-5-E |
Carboxylesterase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5610 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-1.74 |
-7.55 |
1 |
2 |
0 |
37 |
224.306 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
76
-
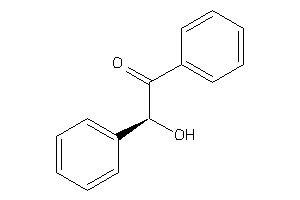
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
7250 |
0.45 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2670 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
6.55 |
-8.14 |
1 |
2 |
0 |
37 |
212.248 |
3 |
↓
|
|
|
|
Analogs
-
3190299
-
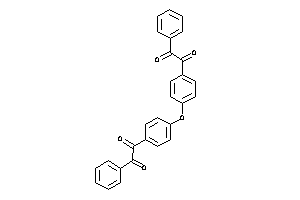
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
3410 |
0.38 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
70 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
2 |
-8.17 |
0 |
4 |
0 |
52 |
270.284 |
5 |
↓
|
|
|
|
Analogs
-
36767369
-
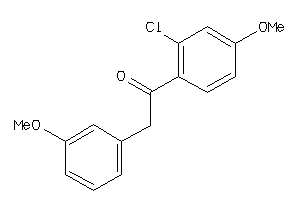
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
3311 |
0.37 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
2.19 |
-10.45 |
0 |
4 |
0 |
52 |
304.729 |
5 |
↓
|
|
|
|
Analogs
-
5363904
-
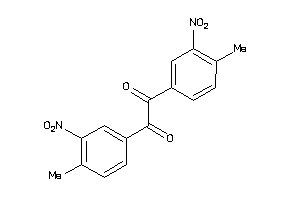
-
2392956
-
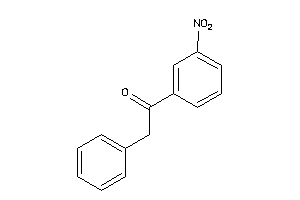
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
2.64 |
-10.46 |
0 |
0 |
0 |
79 |
269.256 |
5 |
↓
|
|
|
|
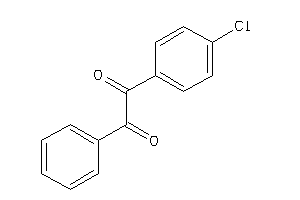
Analogs
-
2170331
-
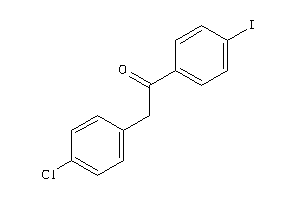
-
2393002
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
182 |
0.52 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
28 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
1.64 |
-4.76 |
0 |
2 |
0 |
34 |
279.122 |
3 |
↓
|
|
|
|
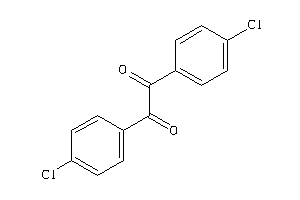
Analogs
-
2393002
-

-
1555402
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.68 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
30 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
1.79 |
-6.01 |
0 |
2 |
0 |
34 |
244.677 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
78 |
0.55 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
21 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
1.36 |
-7.68 |
0 |
2 |
0 |
34 |
303.155 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
160 |
0.53 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
23 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
2.11 |
-5.81 |
0 |
2 |
0 |
34 |
258.704 |
3 |
↓
|
|
|
|
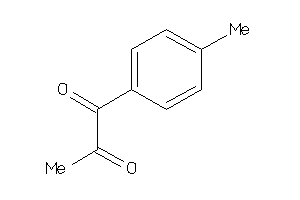
Analogs
-
39234682
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5270 |
0.67 |
Binding ≤ 10μM |
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1840 |
0.73 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
4.9 |
-4.45 |
0 |
2 |
0 |
34 |
148.161 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
231 |
0.52 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
170 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
3.41 |
-5.34 |
0 |
2 |
0 |
34 |
246.212 |
3 |
↓
|
|
|
|
Analogs
-
3190299
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
175 |
0.53 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
7.08 |
-7.89 |
0 |
3 |
0 |
43 |
240.258 |
4 |
↓
|
|
|
|
Analogs
-
2392958
-
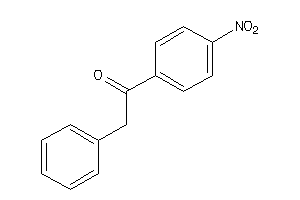
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
215 |
0.49 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
31 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
2.45 |
-7.84 |
0 |
5 |
0 |
79 |
255.229 |
4 |
↓
|
|
|
|
Analogs
-
3200262
-
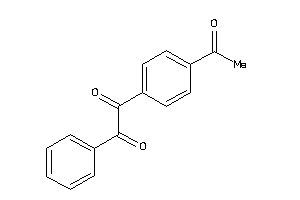
-
39234682
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
537 |
0.49 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
60 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
2.57 |
-7.37 |
0 |
2 |
0 |
34 |
238.286 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-12-E |
Acetylcholinesterase (cluster #12 Of 12), Eukaryotic |
Eukaryotes |
163 |
0.79 |
Binding ≤ 10μM
|
|
CHLE-5-E |
Butyrylcholinesterase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
268 |
0.77 |
Binding ≤ 10μM
|
|
EST1-3-E |
Carboxylesterase (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
84 |
0.83 |
Binding ≤ 10μM
|
|
EST2-4-E |
Carboxylesterase 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
57 |
0.85 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
4.81 |
-1.46 |
0 |
2 |
0 |
34 |
245.876 |
0 |
↓
|
|
|
|
Analogs
-
1710982
-
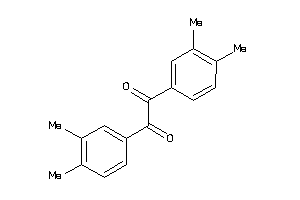
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
99 |
0.54 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
92 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
2.55 |
-7.71 |
0 |
2 |
0 |
34 |
238.286 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
1698 |
0.31 |
Binding ≤ 10μM
|
|
EST2-3-E |
Carboxylesterase 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
380 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.24 |
10.49 |
-1.1 |
0 |
2 |
0 |
34 |
472.407 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
9400 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
2.93 |
-6.56 |
1 |
3 |
0 |
50 |
216.023 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
610 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
-0.79 |
-13.31 |
1 |
4 |
0 |
51 |
286.718 |
3 |
↓
|
|
|
|
Analogs
-
42101101
-

-
42101255
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-5-E |
Acetylcholinesterase (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
3280 |
0.21 |
Binding ≤ 10μM
|
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
2.24 |
-21.78 |
0 |
6 |
0 |
78 |
486.527 |
6 |
↓
|
|
|
|
Analogs
-
4123199
-
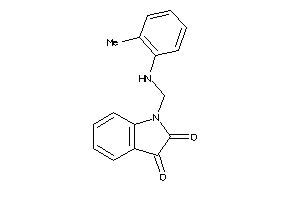
-
4123200
-
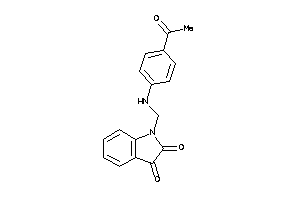
-
4797648
-
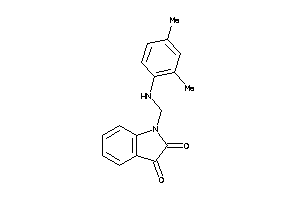
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2650 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
-0.32 |
-12.96 |
1 |
4 |
0 |
51 |
266.3 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
38409987
-
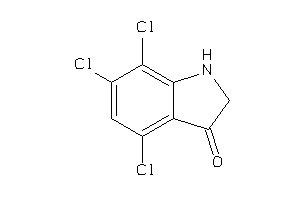
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
590 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
-1.11 |
-7.89 |
1 |
3 |
0 |
50 |
216.023 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2630 |
0.71 |
Binding ≤ 10μM
|
|
EST1-6-E |
Carboxylesterase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
5240 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
4.57 |
-9.29 |
0 |
2 |
0 |
34 |
146.145 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-5-E |
Acetylcholinesterase (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
903 |
0.42 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3100 |
0.39 |
Binding ≤ 10μM
|
|
EST1-4-E |
Carboxylesterase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
570 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
2.82 |
-15.98 |
0 |
8 |
0 |
125 |
272.172 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-4-E |
Carboxylesterase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
1.53 |
-12.28 |
0 |
2 |
0 |
34 |
232.238 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-1-E |
Carboxylesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
5400 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.71 |
4.16 |
-11.27 |
0 |
4 |
0 |
60 |
210.192 |
0 |
↓
|
|