|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C1R-1-E |
Complement C1r (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.46 |
Binding ≤ 10μM |
|
C1S-1-E |
Complement C1s (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1250 |
0.46 |
Binding ≤ 10μM |
|
KLK1-1-E |
Kallikrein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9890 |
0.39 |
Binding ≤ 10μM |
|
PLMN-1-E |
Plasminogen (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6680 |
0.40 |
Binding ≤ 10μM |
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
510 |
0.49 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
1.15 |
-8.83 |
0 |
3 |
0 |
43 |
349.127 |
1 |
↓
|
|
|
|
Analogs
-
4426033
-
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
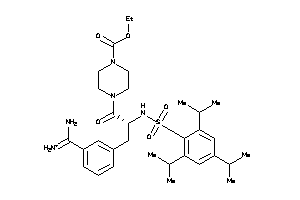
4.30 |
10.74 |
-41.24 |
5 |
10 |
1 |
148 |
614.833 |
12 |
↓
|
|
|
|
Analogs
-
4426028
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.19 |
Binding ≤ 10μM
|
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.23 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
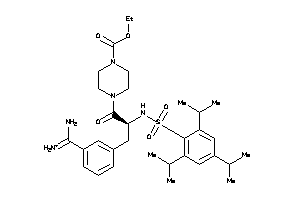
4.30 |
10.52 |
-48.64 |
5 |
10 |
1 |
148 |
614.833 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
-0.38 |
-8.04 |
0 |
7 |
0 |
86 |
350.377 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-1-E |
Plasminogen (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
1.2 |
-13.6 |
3 |
6 |
0 |
100 |
300.266 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.51 |
2.18 |
-57.57 |
2 |
6 |
-1 |
103 |
299.258 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C1R-1-E |
Complement C1r (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1040 |
0.32 |
Binding ≤ 10μM |
|
C1S-1-E |
Complement C1s (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.37 |
Binding ≤ 10μM |
|
KLK1-1-E |
Kallikrein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM |
|
PLMN-1-E |
Plasminogen (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.30 |
Binding ≤ 10μM
|
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Binding ≤ 10μM |
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
7.87 |
-80.13 |
9 |
7 |
2 |
142 |
349.394 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
4.78 |
-43.04 |
5 |
8 |
1 |
127 |
453.544 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.97 |
-4.39 |
-75.96 |
5 |
10 |
0 |
154 |
471.521 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.25 |
Binding ≤ 10μM
|
|
PROC-1-E |
Vitamin K-dependent Protein C (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.24 |
Binding ≤ 10μM
|
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.49 |
-40.89 |
5 |
7 |
1 |
118 |
423.518 |
6 |
↓
|
|