|
|
Analogs
-
2369677
-
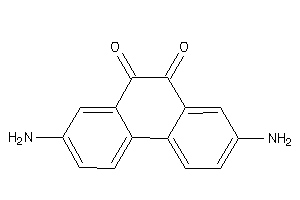
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
5.46 |
-11.78 |
2 |
3 |
0 |
60 |
223.231 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
2.12 |
-15.09 |
0 |
5 |
0 |
79 |
253.213 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTPRC-3-E |
Leukocyte Common Antigen (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM
|
|
Z81338-1-O |
T-cells (cluster #1 Of 3), Other |
Other |
1300 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
2.09 |
-10.2 |
0 |
5 |
0 |
79 |
253.213 |
1 |
↓
|
|
|
|
Analogs
-
2163434
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-2-E |
Dual Specificity Protein Phosphatase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.57 |
Binding ≤ 10μM
|
|
DUS6-1-E |
Dual Specificity Protein Phosphatase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1350 |
0.55 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
270 |
0.61 |
Binding ≤ 10μM
|
|
PTPRC-3-E |
Leukocyte Common Antigen (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
370 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
1.93 |
-10.73 |
0 |
3 |
0 |
43 |
202.209 |
2 |
↓
|
|
|
|
Analogs
-
2163434
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTPRC-3-E |
Leukocyte Common Antigen (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
2.38 |
-10.42 |
0 |
3 |
0 |
43 |
214.22 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
8.51 |
-21.23 |
1 |
4 |
0 |
63 |
307.349 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
7.58 |
-11.46 |
0 |
2 |
0 |
34 |
208.216 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
6.09 |
-12.34 |
0 |
2 |
0 |
34 |
158.156 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTPRC-3-E |
Leukocyte Common Antigen (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.53 |
Binding ≤ 10μM
|
|
Z81338-1-O |
T-cells (cluster #1 Of 3), Other |
Other |
600 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
8.2 |
-10.88 |
0 |
2 |
0 |
34 |
287.112 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.19 |
Binding ≤ 10μM
|
|
DUS3-2-E |
Dual Specificity Protein Phosphatase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.23 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2200 |
0.20 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.20 |
Binding ≤ 10μM
|
|
PTPRC-3-E |
Leukocyte Common Antigen (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.52 |
8.73 |
-53.3 |
0 |
10 |
-1 |
134 |
617.776 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.27 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.20 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
460 |
0.22 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.22 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
12.53 |
-51.54 |
0 |
8 |
-1 |
106 |
611.146 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
780 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.26 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.20 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.21 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
610 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
12.04 |
-51.46 |
0 |
8 |
-1 |
106 |
576.701 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
71 |
0.25 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.19 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
590 |
0.22 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
12.62 |
-50.63 |
0 |
8 |
-1 |
106 |
611.146 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-1-E |
Dual Specificity Protein Phosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.22 |
Binding ≤ 10μM
|
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
74 |
0.25 |
Binding ≤ 10μM
|
|
MPIP1-2-E |
Dual Specificity Phosphatase Cdc25A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.19 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
420 |
0.22 |
Binding ≤ 10μM
|
|
PTN7-1-E |
Protein-tyrosine Phosphatase LC-PTP (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
12.21 |
-50.97 |
0 |
8 |
-1 |
106 |
594.691 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9700 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
1.39 |
-62.88 |
1 |
6 |
-1 |
95 |
470.526 |
8 |
↓
|
|