|
|
Analogs
-
3072905
-
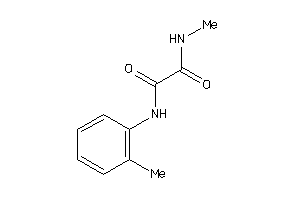
-
8737440
-
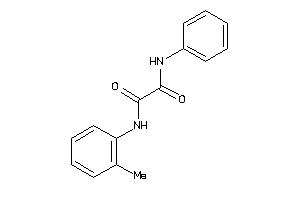
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGM-3-E |
Muscle Glycogen Phosphorylase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5400 |
0.37 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
6.55 |
-6.66 |
2 |
4 |
0 |
58 |
268.316 |
3 |
↓
|
|
|
|
Analogs
-
3978828
-
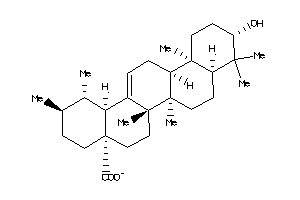
-
3978829
-
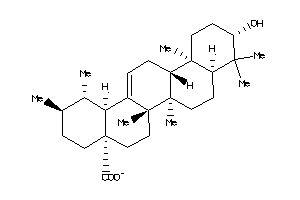
-
4273370
-
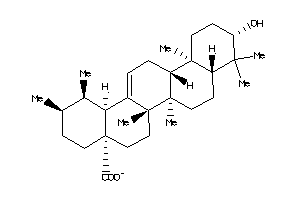
-
4273371
-
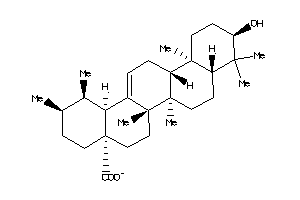
-
4273372
-
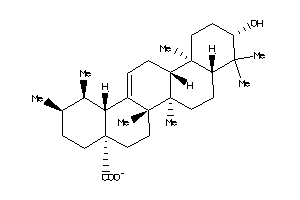
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
DPOLB-2-E |
DNA Polymerase Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.22 |
Binding ≤ 10μM
|
|
PA21B-3-E |
Phospholipase A2 Group 1B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.23 |
Binding ≤ 10μM
|
|
PA2A-1-E |
Phospholipase A2 Isozyme PLA-A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GA-3-E |
Phospholipase A2, Membrane Associated (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GD-2-E |
Group IID Secretory Phospholipase A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GE-2-E |
Group IIE Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GF-2-E |
Group IIF Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.23 |
Binding ≤ 10μM
|
|
PTN2-2-E |
T-cell Protein-tyrosine Phosphatase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6700 |
0.22 |
Binding ≤ 10μM |
|
PYGM-1-E |
Muscle Glycogen Phosphorylase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.21 |
Binding ≤ 10μM |
|
Q7T3S7-1-E |
Phospholipase A2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1430 |
0.25 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-4-O |
Trypanosoma Brucei (cluster #4 Of 6), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50420-3-O |
Trypanosoma Brucei Brucei (cluster #3 Of 7), Other |
Other |
2200 |
0.24 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50472-2-O |
Toxoplasma Gondii (cluster #2 Of 4), Other |
Other |
1000 |
0.25 |
Functional ≤ 10μM
|
|
Z50607-8-O |
Human Immunodeficiency Virus 1 (cluster #8 Of 10), Other |
Other |
1800 |
0.24 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
4400 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
12.05 |
-49.78 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.79 |
10.29 |
-5.45 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGM-2-E |
Muscle Glycogen Phosphorylase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3200 |
0.35 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
-0.98 |
-8.13 |
0 |
3 |
0 |
38 |
306.752 |
2 |
↓
|
|
|
|
Analogs
-
36889913
-
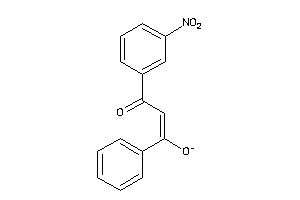
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGM-3-E |
Muscle Glycogen Phosphorylase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.34 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
9.12 |
-39.17 |
0 |
6 |
-1 |
95 |
298.274 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
3.28 |
-29.1 |
0 |
6 |
0 |
89 |
299.282 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGM-3-E |
Muscle Glycogen Phosphorylase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
-1.88 |
-5.94 |
2 |
4 |
0 |
58 |
288.734 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
2585429
-
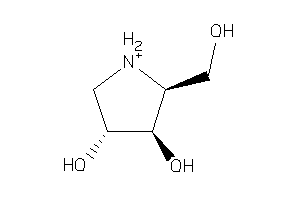
-
15657755
-
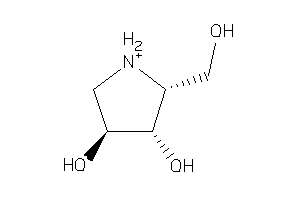
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WC34-1-E |
Alpha-glucosidase II Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9700 |
0.78 |
Binding ≤ 10μM
|
|
D3ZAN3-1-E |
Alpha-glucosidase II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9700 |
0.78 |
Binding ≤ 10μM
|
|
GDE-1-E |
Glycogen Debranching Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.79 |
Binding ≤ 10μM
|
|
PYGL-2-E |
Liver Glycogen Phosphorylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
370 |
1.00 |
Binding ≤ 10μM
|
|
PYGM-4-E |
Muscle Glycogen Phosphorylase (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
396 |
1.00 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5800 |
0.81 |
Binding ≤ 10μM
|
|
AMYG-1-F |
Glucoamylase, Intracellular Sporulation-specific (cluster #1 Of 1), Fungal |
Fungi |
150 |
1.06 |
Binding ≤ 10μM
|
|
MAL62-2-F |
Alpha-glucosidase MAL62 (cluster #2 Of 2), Fungal |
Fungi |
840 |
0.95 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.98 |
-7.72 |
-35.36 |
5 |
4 |
1 |
77 |
134.155 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.98 |
-9.1 |
-6.49 |
4 |
4 |
0 |
73 |
133.147 |
1 |
↓
|
|
|
|
Analogs
-
12502895
-
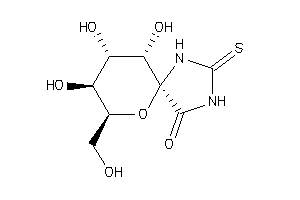
-
12502898
-
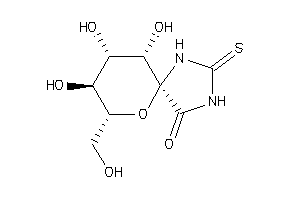
-
3872695
-
-
3872694
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
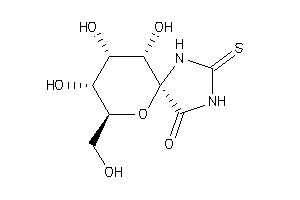
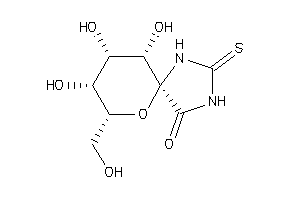
(2R,3S,4S,5R,6S)-3,4,5-Trihydroxy-2-hydroxymethyl-7,9-diaza-1-oxa-spiro[4,5]decane-10-one-8-thione
(2R,3S,4S,5R,6S)-3,4,5-Trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGB-1-E |
Brain Glycogen Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
PYGL-2-E |
Liver Glycogen Phosphorylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5100 |
0.44 |
Binding ≤ 10μM
|
|
PYGM-4-E |
Muscle Glycogen Phosphorylase (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5100 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.78 |
-16.4 |
-14.06 |
6 |
8 |
0 |
131 |
264.259 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1650 |
0.23 |
Binding ≤ 10μM
|
|
PTN2-2-E |
T-cell Protein-tyrosine Phosphatase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5990 |
0.21 |
Binding ≤ 10μM
|
|
PYGM-1-E |
Muscle Glycogen Phosphorylase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.20 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.17 |
13.92 |
-54.4 |
1 |
4 |
-1 |
69 |
477.713 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.17 |
14.08 |
-75.61 |
2 |
4 |
0 |
70 |
478.721 |
1 |
↓
|
|
|
|
|