|
|
Analogs
-
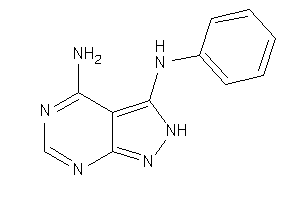
34333011
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Binding ≤ 10μM |
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.47 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.45 |
Binding ≤ 10μM
|
|
SGK1-1-E |
Serine/threonine-protein Kinase Sgk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
5.16 |
-9.26 |
4 |
6 |
0 |
93 |
244.233 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.30 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.30 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.30 |
Binding ≤ 10μM
|
|
DYRK2-1-E |
Dual-specificity Tyrosine-phosphorylation Regulated Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM |
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
40 |
0.38 |
Binding ≤ 10μM
|
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2600 |
0.29 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.28 |
Binding ≤ 10μM
|
|
MP2K6-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.39 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
ROCK2-1-E |
Rho-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
SGK1-1-E |
Serine/threonine-protein Kinase Sgk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
10.69 |
-11.56 |
2 |
6 |
0 |
90 |
372.457 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.46 |
11.16 |
-40.43 |
3 |
6 |
1 |
92 |
373.465 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.46 |
11.46 |
-42.48 |
3 |
6 |
1 |
92 |
373.465 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-3-E |
Cyclin A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2380 |
0.49 |
Binding ≤ 10μM
|
|
CCNA2-3-E |
Cyclin A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2380 |
0.49 |
Binding ≤ 10μM
|
|
CDK2-5-E |
Cyclin-dependent Kinase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2380 |
0.49 |
Binding ≤ 10μM
|
|
CSK21-3-E |
Casein Kinase II Alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5790 |
0.46 |
Binding ≤ 10μM
|
|
CSK22-2-E |
Casein Kinase II Alpha (prime) (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5790 |
0.46 |
Binding ≤ 10μM
|
|
CSK2B-2-E |
Casein Kinase II Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5790 |
0.46 |
Binding ≤ 10μM
|
|
DYR1A-2-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.61 |
Binding ≤ 10μM
|
|
KPBB-2-E |
Phosphorylase Kinase Beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7040 |
0.45 |
Binding ≤ 10μM
|
|
PHKG1-2-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7040 |
0.45 |
Binding ≤ 10μM
|
|
PHKG2-2-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7040 |
0.45 |
Binding ≤ 10μM
|
|
SGK1-2-E |
Serine/threonine-protein Kinase Sgk1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3570 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
8.43 |
-5.86 |
1 |
3 |
0 |
32 |
476.792 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4430 |
0.21 |
Binding ≤ 10μM
|
|
AKT2-2-E |
Serine/threonine-protein Kinase AKT2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4430 |
0.21 |
Binding ≤ 10μM
|
|
AKT3-2-E |
Serine/threonine-protein Kinase AKT3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4430 |
0.21 |
Binding ≤ 10μM
|
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8090 |
0.20 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
SGK1-1-E |
Serine/threonine-protein Kinase Sgk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1320 |
0.24 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
7.49 |
-24.17 |
4 |
6 |
0 |
98 |
465.553 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGK1-1-E |
Serine/threonine-protein Kinase Sgk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
580 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
14.67 |
-59.57 |
1 |
4 |
-1 |
69 |
381.455 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGK1-1-E |
Serine/threonine-protein Kinase Sgk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
9.3 |
-44.99 |
1 |
6 |
-1 |
81 |
337.366 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.35 |
9.3 |
-11.56 |
2 |
6 |
0 |
83 |
338.374 |
3 |
↓
|
|