|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6600 |
0.38 |
Binding ≤ 10μM |
|
TRPV1-3-E |
Vanilloid Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.44 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
5.89 |
-7.35 |
2 |
3 |
0 |
49 |
263.381 |
8 |
↓
|
|
|
|
Analogs
-
3883031
-

-
365745
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
308 |
0.61 |
Binding ≤ 10μM |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
0.76 |
-9.36 |
0 |
2 |
0 |
26 |
195.221 |
0 |
↓
|
|
|
|
Analogs
-
5934388
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
10 |
1.12 |
Binding ≤ 10μM |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10 |
1.12 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
1.94 |
-5.44 |
0 |
1 |
0 |
23 |
196.047 |
1 |
↓
|
|
|
|
Analogs
-
5934150
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
10 |
1.12 |
Binding ≤ 10μM |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10 |
1.12 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
1.94 |
-5.42 |
0 |
1 |
0 |
23 |
196.047 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1100 |
1.67 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.60 |
2.72 |
-6.89 |
0 |
1 |
0 |
17 |
136.976 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
39 |
1.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
3.79 |
-5.34 |
0 |
2 |
0 |
26 |
167.002 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1900 |
0.73 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
2.41 |
-11.55 |
0 |
2 |
0 |
34 |
154.209 |
6 |
↓
|
|
|
|
Analogs
-
5735314
-
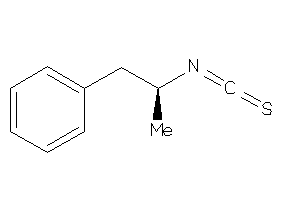
-
5751594
-
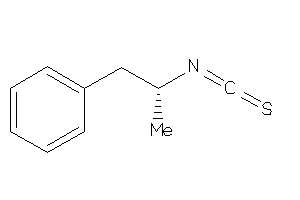
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM |
|
CP2E1-1-E |
Cytochrome P450 2E1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9980 |
0.64 |
ADME/T ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
9800 |
0.64 |
Functional ≤ 10μM |
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
9700 |
0.64 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
5.69 |
-2.35 |
0 |
1 |
0 |
12 |
163.245 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1900 |
1.33 |
Binding ≤ 10μM |
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5813 |
1.22 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
3.02 |
-27.34 |
1 |
1 |
1 |
14 |
100.166 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.25 |
2.45 |
-1.96 |
0 |
1 |
0 |
12 |
99.158 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5000 |
1.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
3.38 |
-5.99 |
0 |
1 |
0 |
17 |
84.118 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
1580 |
1.02 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.98 |
-1.44 |
-55.14 |
3 |
3 |
1 |
62 |
156.252 |
3 |
↓
|
|
|
|
Analogs
-
13523661
-
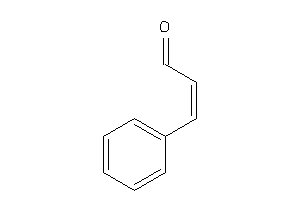
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
6500 |
0.73 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
5.49 |
-7.64 |
0 |
1 |
0 |
17 |
132.162 |
2 |
↓
|
|
|
|
Analogs
-
4097409
-
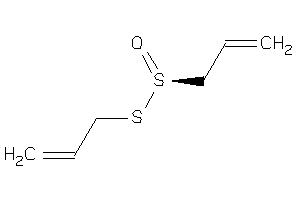
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-3-E |
Cathepsin B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8600 |
0.79 |
Binding ≤ 10μM
|
|
CATL1-2-E |
Cathepsin L (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9300 |
0.78 |
Binding ≤ 10μM
|
|
Q95PM0-3-E |
Rhodesain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5310 |
0.82 |
Binding ≤ 10μM
|
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
1320 |
0.91 |
Binding ≤ 10μM |
|
Z50425-19-O |
Plasmodium Falciparum (cluster #19 Of 22), Other |
Other |
5210 |
0.82 |
Functional ≤ 10μM
|
|
Z80156-11-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #11 Of 12), Other |
Other |
5500 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
-0.02 |
-9.12 |
0 |
1 |
0 |
17 |
162.279 |
5 |
↓
|
|
|
|
Analogs
-
1530846
-
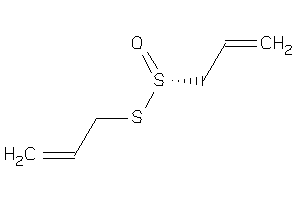
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-3-E |
Cathepsin B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
8600 |
0.79 |
Binding ≤ 10μM
|
|
CATL1-2-E |
Cathepsin L (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9300 |
0.78 |
Binding ≤ 10μM
|
|
Q95PM0-3-E |
Rhodesain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5310 |
0.82 |
Binding ≤ 10μM
|
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
1320 |
0.91 |
Binding ≤ 10μM |
|
Z50425-19-O |
Plasmodium Falciparum (cluster #19 Of 22), Other |
Other |
5210 |
0.82 |
Functional ≤ 10μM
|
|
Z80156-11-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #11 Of 12), Other |
Other |
5500 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
-0.26 |
-9.78 |
0 |
1 |
0 |
17 |
162.279 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
800 |
0.57 |
Binding ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.57 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
6.08 |
-9.65 |
0 |
3 |
0 |
39 |
201.225 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5600 |
0.32 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
13.63 |
-48.4 |
0 |
3 |
-1 |
57 |
315.433 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
319 |
0.91 |
Binding ≤ 10μM |
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
7400 |
0.72 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
4.91 |
-2.94 |
0 |
1 |
0 |
12 |
149.218 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.01 |
5.47 |
-29.74 |
1 |
1 |
1 |
14 |
150.226 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
800 |
2.13 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.78 |
1.73 |
-6.54 |
0 |
1 |
0 |
17 |
56.064 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
30 |
1.05 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
5.62 |
-8.14 |
0 |
1 |
0 |
17 |
154.596 |
2 |
↓
|
|
|
|
Analogs
-
7641734
-
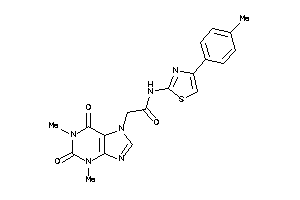
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
-0.65 |
-21.82 |
1 |
9 |
0 |
103 |
396.432 |
4 |
↓
|
|
|
|
Analogs
-
2147303
-
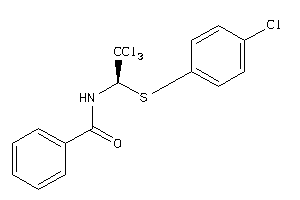
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.14 |
-0.95 |
-4.86 |
1 |
2 |
0 |
29 |
409.165 |
5 |
↓
|
|
|
|
Analogs
-
2147303
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.14 |
-0.87 |
-5.25 |
1 |
2 |
0 |
29 |
409.165 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
15 |
0.58 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.2 |
-7.64 |
1 |
3 |
0 |
46 |
361.077 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
15 |
0.58 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.26 |
-9.14 |
1 |
3 |
0 |
46 |
361.077 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
8.72 |
-23.58 |
1 |
10 |
0 |
113 |
426.458 |
5 |
↓
|
|
|
|
Analogs
-
4938919
-
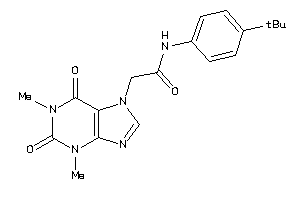
-
5165262
-
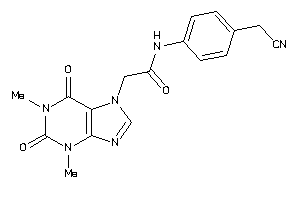
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7500 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
0.85 |
-19.35 |
1 |
8 |
0 |
90 |
355.398 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.38 |
1.13 |
-54.9 |
2 |
8 |
1 |
92 |
356.406 |
4 |
↓
|
|
|
|
Analogs
-
2147305
-
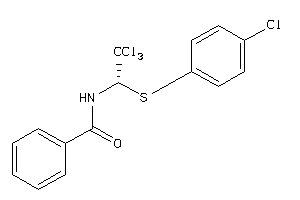
-
3138918
-
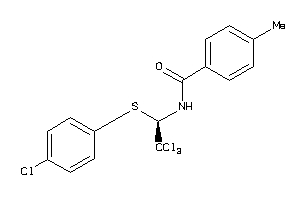
-
3138919
-
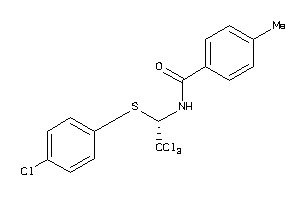
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
66 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
7.74 |
-4.11 |
1 |
2 |
0 |
33 |
395.138 |
5 |
↓
|
|
|
|
Analogs
-
3138918
-

-
3138919
-

-
2147303
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
66 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
7.66 |
-4.12 |
1 |
2 |
0 |
33 |
395.138 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.54 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
1.43 |
-3.09 |
0 |
4 |
0 |
59 |
174.159 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
10.04 |
-21.14 |
1 |
9 |
0 |
104 |
475.328 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.40 |
10.53 |
-60.32 |
2 |
9 |
1 |
105 |
476.336 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
115 |
0.40 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
-1.23 |
-5.88 |
1 |
3 |
0 |
38 |
425.164 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
115 |
0.40 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
-1.16 |
-6.22 |
1 |
3 |
0 |
38 |
425.164 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
35 |
0.42 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.65 |
-0.75 |
-7.55 |
1 |
5 |
0 |
74 |
440.135 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
35 |
0.42 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.65 |
-0.68 |
-7.97 |
1 |
5 |
0 |
74 |
440.135 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
51 |
0.44 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.50 |
10.88 |
-4.33 |
1 |
2 |
0 |
29 |
474.034 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
51 |
0.44 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.50 |
10.92 |
-4.73 |
1 |
2 |
0 |
29 |
474.034 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
11.08 |
-7.99 |
1 |
5 |
0 |
75 |
419.717 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
11.12 |
-8.23 |
1 |
5 |
0 |
75 |
419.717 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.98 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
9.09 |
-2.27 |
0 |
0 |
0 |
0 |
178.347 |
6 |
↓
|
|
|
|
Analogs
-
2163483
-
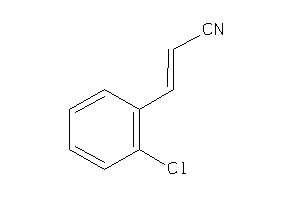
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.97 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
3.87 |
-6.09 |
0 |
2 |
0 |
48 |
188.617 |
1 |
↓
|
|
|
|
Analogs
-
9227019
-
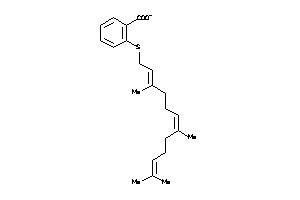
-
13161992
-
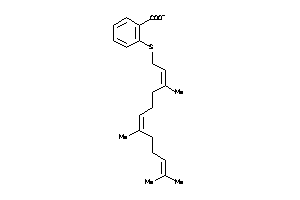
-
13161993
-
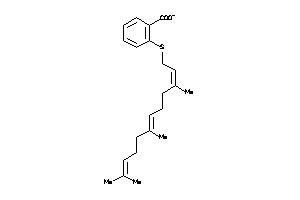
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ICMT-2-E |
Isoprenylcysteine Carboxyl Methyltransferase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.31 |
Binding ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Functional ≤ 10μM |
|
Z80130-1-O |
EJ (Bladder Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
7500 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.26 |
15.85 |
-52.19 |
0 |
2 |
-1 |
40 |
357.539 |
10 |
↓
|
|
|
|
|
|
|
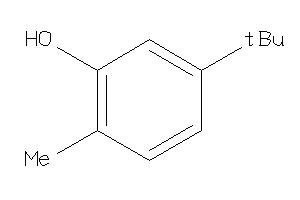
Analogs
-
2582758
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4000 |
0.69 |
Binding ≤ 10μM
|
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7000 |
0.66 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
4.3 |
-3.43 |
1 |
1 |
0 |
20 |
150.221 |
1 |
↓
|
|
|
|
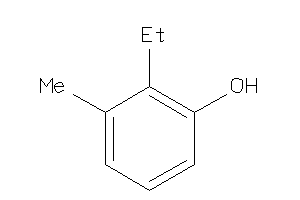
Analogs
-
5688529
-
-
5688577
-
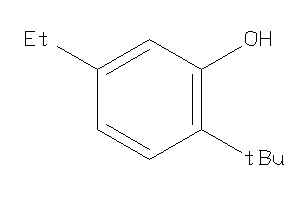
-
5782381
-
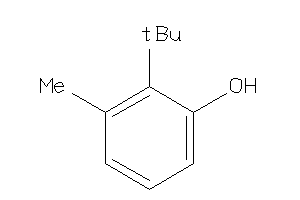
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-1-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.66 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
4.34 |
-3.43 |
1 |
1 |
0 |
20 |
150.221 |
1 |
↓
|
|
|
|
Analogs
-
1531088
-
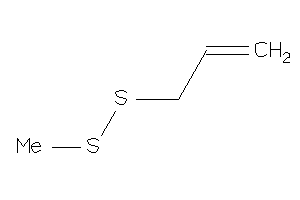
-
32163213
-
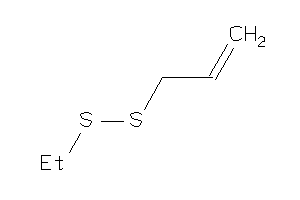
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-5-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7550 |
0.90 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
7.01 |
-2.24 |
0 |
0 |
0 |
0 |
146.28 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.32 |
Binding ≤ 10μM
|
|
TRPV1-2-E |
Vanilloid Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
34 |
0.40 |
Binding ≤ 10μM
|
|
TRPA1-2-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4500 |
0.29 |
Functional ≤ 10μM
|
|
TRPV1-1-E |
Vanilloid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.72 |
9.65 |
-12.18 |
1 |
5 |
0 |
48 |
372.9 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2580 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
8.77 |
-10.31 |
0 |
2 |
0 |
34 |
232.323 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
980 |
0.37 |
Binding ≤ 10μM
|
|
TRPA1-3-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.24 |
14.88 |
-48.79 |
0 |
5 |
-1 |
86 |
326.457 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.24 |
12.9 |
-11.26 |
1 |
5 |
0 |
83 |
327.465 |
16 |
↓
|
|
|
|
Analogs
-
8552978
-
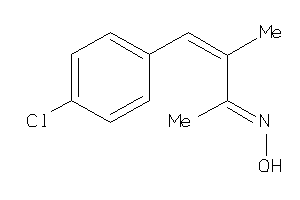
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
4500 |
0.53 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
0.51 |
-5.49 |
1 |
2 |
0 |
32 |
209.676 |
2 |
↓
|
|