|
|
Analogs
-
4468952
-
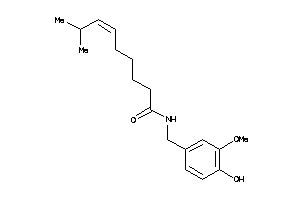
-
33845128
-
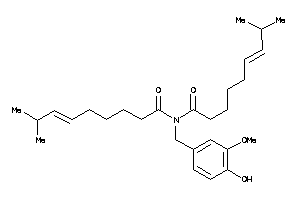
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
6.6 |
-9.89 |
2 |
4 |
0 |
59 |
305.418 |
9 |
↓
|
|
|
|
Analogs
-
34977778
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPV1-1-E |
Vanilloid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.70 |
Functional ≤ 10μM
|
|
TRPV2-2-E |
Transient Receptor Potential Cation Channel Subfamily V Member 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.70 |
Functional ≤ 10μM
|
|
TRPV4-2-E |
Transient Receptor Potential Cation Channel Subfamily V Member 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
1.76 |
-39.64 |
3 |
5 |
1 |
64 |
207.257 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.31 |
1.48 |
-13.79 |
2 |
5 |
0 |
62 |
206.249 |
1 |
↓
|
|
|
|
Analogs
-
5757941
-
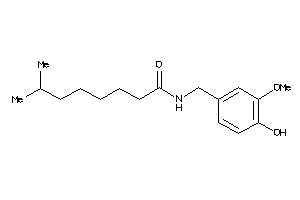
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPV1-1-E |
Vanilloid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.43 |
Functional ≤ 10μM
|
|
TRPV2-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
190 |
0.43 |
Functional ≤ 10μM
|
|
TRPV4-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
190 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
6.63 |
-9.86 |
2 |
4 |
0 |
59 |
307.434 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPV1-1-E |
Vanilloid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
TRPV1-1-E |
Vanilloid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Functional ≤ 10μM
|
|
TRPV2-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.37 |
Functional ≤ 10μM
|
|
TRPV4-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
7.82 |
-45.77 |
2 |
5 |
-1 |
72 |
422.571 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.16 |
7.88 |
-45.18 |
2 |
5 |
-1 |
74 |
422.571 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.12 |
7.63 |
-16.83 |
3 |
5 |
0 |
70 |
423.579 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.74 |
-18.05 |
3 |
4 |
0 |
56 |
376.909 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.37 |
Binding ≤ 10μM |
|
CNR2-3-E |
Cannabinoid CB2 Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
880 |
0.37 |
Binding ≤ 10μM |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
700 |
0.37 |
Functional ≤ 10μM |
|
TRPM8-1-E |
Transient Receptor Potential Cation Channel Subfamily M Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.41 |
Functional ≤ 10μM |
|
TRPV1-2-E |
Vanilloid Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Functional ≤ 10μM |
|
TRPV2-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.84 |
0.36 |
-4.27 |
2 |
2 |
0 |
40 |
316.485 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
7.21 |
-11.32 |
3 |
4 |
0 |
55 |
324.49 |
11 |
↓
|
|
Ref
Reference (pH 7)
|
4.49 |
7.31 |
-12.83 |
3 |
4 |
0 |
54 |
324.49 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.53 |
6.94 |
-34.66 |
3 |
4 |
1 |
55 |
325.498 |
11 |
↓
|
|
|
|
Analogs
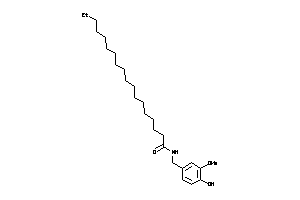
-
43273992
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
-1.49 |
-9.46 |
2 |
4 |
0 |
58 |
293.407 |
10 |
↓
|
|
|
|
Analogs
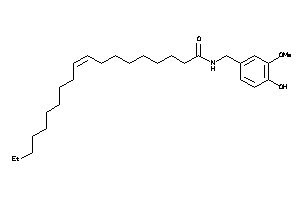
-
64858452
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPV1-1-E |
Vanilloid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.32 |
Functional ≤ 10μM
|
|
TRPV2-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
170 |
0.32 |
Functional ≤ 10μM
|
|
TRPV4-1-E |
Transient Receptor Potential Cation Channel Subfamily V Member 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
170 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.18 |
-0.2 |
-9.68 |
2 |
4 |
0 |
58 |
417.634 |
18 |
↓
|
|