|
|
Analogs
-
6199057
-
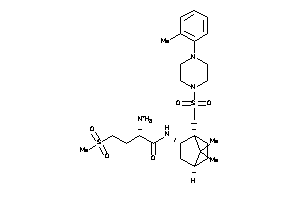
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-9.88 |
-61.78 |
4 |
9 |
1 |
131 |
555.787 |
9 |
↓
|
|
|
|
Analogs
-
38401134
-
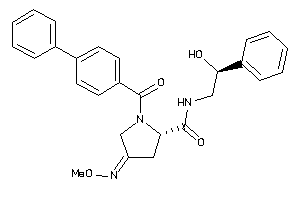
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
135 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
9.93 |
-20.58 |
2 |
7 |
0 |
91 |
471.557 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
6.38 |
-9.94 |
1 |
5 |
0 |
62 |
338.407 |
4 |
↓
|
|
|
|
Analogs
-
1192101
-
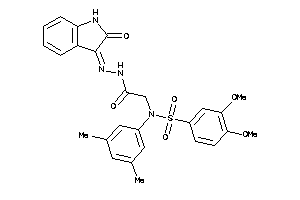
-
16226943
-
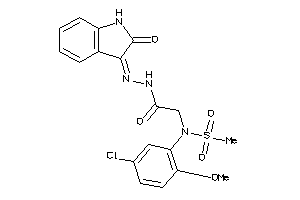
-
16227484
-
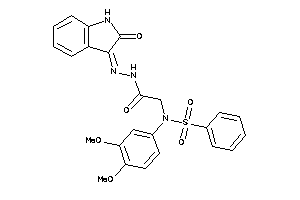
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1410 |
0.23 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.27 |
-23.27 |
2 |
9 |
0 |
121 |
492.557 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.60 |
5.53 |
-56.09 |
1 |
9 |
-1 |
124 |
491.549 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.60 |
5.27 |
-72.79 |
1 |
9 |
-1 |
124 |
491.549 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
V1AR-2-E |
Vasopressin V1a Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Functional ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.69 |
14.99 |
-45.21 |
3 |
6 |
1 |
79 |
499.594 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.69 |
14.56 |
-20.71 |
2 |
6 |
0 |
78 |
498.586 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.69 |
14.57 |
-17.85 |
2 |
6 |
0 |
78 |
498.586 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
10.8 |
-30.29 |
0 |
9 |
0 |
89 |
507.587 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.28 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.27 |
Binding ≤ 10μM |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.27 |
Functional ≤ 10μM |
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.27 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
-1.73 |
-21.51 |
1 |
7 |
0 |
78 |
449.551 |
7 |
↓
|
|
|
|
Analogs
-
1490477
-
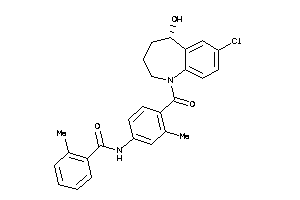
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
-0.59 |
-11.63 |
2 |
5 |
0 |
69 |
448.95 |
3 |
↓
|
|
|
|
Analogs
-
538658
-
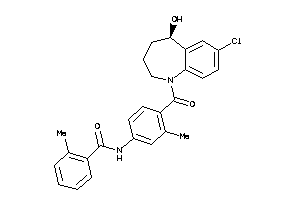
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
10.9 |
-11.46 |
2 |
5 |
0 |
70 |
448.95 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.30 |
Binding ≤ 10μM |
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
285 |
0.22 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.27 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
6.62 |
-25.09 |
3 |
10 |
0 |
139 |
620.511 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.25 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.23 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.71 |
2.49 |
-48 |
2 |
5 |
1 |
53 |
428.556 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.25 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.23 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.71 |
2.46 |
-55.15 |
2 |
5 |
1 |
53 |
428.556 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.34 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
63 |
0.31 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
7.4 |
-18.8 |
0 |
8 |
0 |
90 |
463.563 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.30 |
7.81 |
-48.67 |
1 |
8 |
1 |
91 |
464.571 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
519 |
0.26 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.22 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.36 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
91 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.51 |
1.8 |
-12.67 |
1 |
5 |
0 |
54 |
473.935 |
3 |
↓
|
|
|
|
Analogs
-
43200569
-
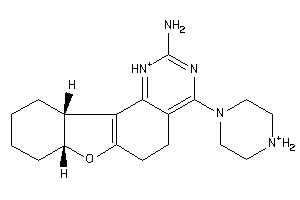
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
8.61 |
-85.28 |
5 |
6 |
2 |
82 |
329.448 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.43 |
6.82 |
-7.53 |
3 |
6 |
0 |
76 |
327.432 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.43 |
8.14 |
-47.33 |
4 |
6 |
1 |
81 |
328.44 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
5.04 |
-24.35 |
1 |
11 |
0 |
126 |
630.119 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A5-1-E |
Glycine Transporter 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.31 |
Binding ≤ 10μM |
|
SC6A9-1-E |
Glycine Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.31 |
Binding ≤ 10μM |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.37 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2460 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.95 |
3.3 |
-14.47 |
0 |
4 |
0 |
39 |
341.414 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.33 |
Functional ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
8.55 |
-15.73 |
1 |
6 |
0 |
76 |
444.94 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4900 |
0.24 |
Functional ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
7.13 |
-17.3 |
2 |
7 |
0 |
99 |
457.939 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.53 |
7.9 |
-59.03 |
1 |
7 |
-1 |
102 |
456.931 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.53 |
7.38 |
-48.24 |
1 |
7 |
-1 |
102 |
456.931 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.31 |
Functional ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3215 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
7.08 |
-26.49 |
2 |
8 |
0 |
108 |
467.547 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.84 |
7.86 |
-76.06 |
1 |
8 |
-1 |
111 |
466.539 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.28 |
Functional ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.23 |
8.62 |
-16.03 |
1 |
6 |
0 |
76 |
444.94 |
7 |
↓
|
|
|
|
Analogs
-
656558
-
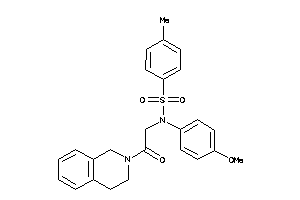
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.31 |
Functional ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
115 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
9.9 |
-22.21 |
0 |
6 |
0 |
67 |
436.533 |
6 |
↓
|
|
|
|
Analogs
-
829925
-
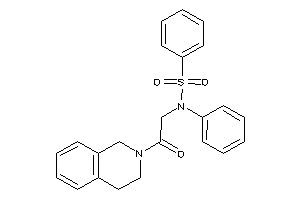
-
1116032
-
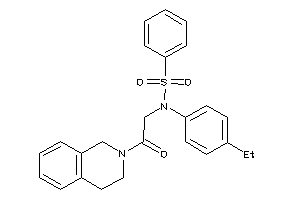
-
9059414
-
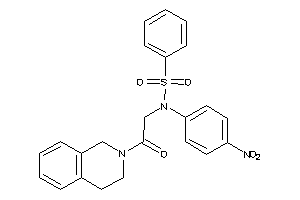
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.30 |
Functional ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
325 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
11.29 |
-20.92 |
0 |
5 |
0 |
58 |
420.534 |
5 |
↓
|
|