|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.17 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.19 |
ADME/T ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 1), Other |
Other |
1 |
0.26 |
Binding ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
4230 |
0.15 |
Functional ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
35 |
0.21 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
960 |
0.17 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
97 |
0.20 |
Functional ≤ 10μM
|
|
Z50614-1-O |
Human T-lymphotropic Virus 1 (cluster #1 Of 1), Other |
Other |
18 |
0.22 |
Functional ≤ 10μM
|
|
Z50658-3-O |
Human Immunodeficiency Virus 2 (cluster #3 Of 4), Other |
Other |
90 |
0.20 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
2 |
0.25 |
Functional ≤ 10μM |
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 3), Other |
Other |
2 |
0.25 |
Functional ≤ 10μM
|
|
Z80076-3-O |
CEM-SS (T-cell Leukemia) (cluster #3 Of 5), Other |
Other |
7 |
0.23 |
Functional ≤ 10μM |
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
5 |
0.24 |
Functional ≤ 10μM |
|
Z80295-1-O |
MT4 (Lymphocytes) (cluster #1 Of 8), Other |
Other |
80 |
0.20 |
Functional ≤ 10μM
|
|
Z80516-1-O |
SUP-T1 (cluster #1 Of 1), Other |
Other |
20 |
0.22 |
Functional ≤ 10μM |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
2 |
0.25 |
Functional ≤ 10μM |
|
Z80897-1-O |
H9 (T-lymphoid Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.22 |
Functional ≤ 10μM |
|
P88142-1-V |
Human Immunodeficiency Virus Type 2 Pol Protein (cluster #1 Of 1), Viral |
Viruses |
1 |
0.26 |
Binding ≤ 10μM
|
|
Q4U254-2-V |
Human Rhinovirus A Protease (cluster #2 Of 3), Viral |
Viruses |
1 |
0.26 |
Binding ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
1 |
0.26 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
-7.47 |
-55.36 |
7 |
11 |
1 |
167 |
671.863 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.16 |
Functional ≤ 10μM
|
|
CP2B6-3-E |
Cytochrome P450 2B6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.16 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.20 |
ADME/T ≤ 10μM
|
|
CP3A5-1-E |
Cytochrome P450 3A5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.19 |
ADME/T ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
2233 |
0.16 |
Functional ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
34 |
0.21 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9664 |
0.14 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
25 |
0.21 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
90 |
0.20 |
Functional ≤ 10μM
|
|
Z50658-3-O |
Human Immunodeficiency Virus 2 (cluster #3 Of 4), Other |
Other |
84 |
0.20 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
38 |
0.21 |
Functional ≤ 10μM
|
|
Z80294-1-O |
MT2 (Lymphocytes) (cluster #1 Of 1), Other |
Other |
10 |
0.22 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
90 |
0.20 |
Functional ≤ 10μM |
|
Z102164-1-O |
Liver Microsomes (cluster #1 Of 1), Other |
Other |
40 |
0.21 |
ADME/T ≤ 10μM
|
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
60 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.51 |
12.91 |
-25.34 |
4 |
11 |
0 |
146 |
720.962 |
18 |
↓
|
|
|
|
Analogs
-
14096305
-
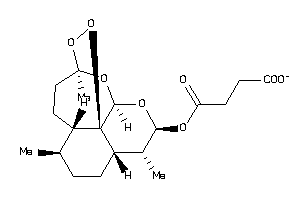
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
-1.53 |
-50.29 |
0 |
8 |
-1 |
103 |
383.417 |
5 |
↓
|
|
|
|
Analogs
-
8214489
-
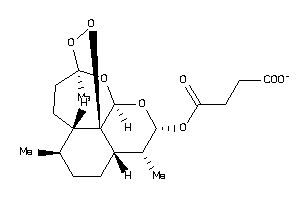
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.73 |
-53.19 |
0 |
8 |
-1 |
103 |
383.417 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
10.21 |
-76.51 |
2 |
6 |
2 |
41 |
537.539 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.60 |
9.81 |
-38.25 |
1 |
6 |
1 |
40 |
536.531 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.60 |
12.59 |
-154.37 |
3 |
6 |
3 |
42 |
538.547 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.89 |
10.56 |
-92.66 |
4 |
7 |
2 |
76 |
520.077 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.89 |
11.26 |
-34.85 |
3 |
7 |
1 |
79 |
519.069 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.89 |
10.95 |
-151.63 |
5 |
7 |
3 |
77 |
521.085 |
7 |
↓
|
|
|
|
Analogs
-
3874185
-
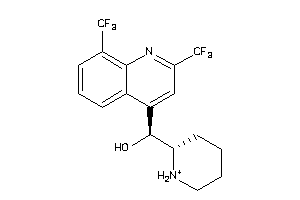
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1221 |
0.32 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
124 |
0.37 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.7 |
-52.23 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6553 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5250 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.35 |
-59.95 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.24 |
5.78 |
-11.48 |
2 |
3 |
0 |
45 |
378.316 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
255 |
0.36 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
61 |
0.39 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.35 |
-59.95 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.24 |
5.69 |
-9.16 |
2 |
3 |
0 |
45 |
378.316 |
4 |
↓
|
|
|
|
Analogs
-
537964
-
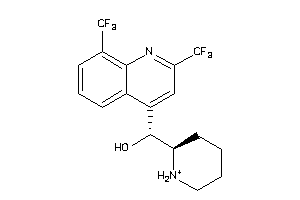
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8607 |
0.27 |
Binding ≤ 10μM
|
|
AA2AR-2-E |
Adenosine A2a Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2534 |
0.30 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5623 |
0.28 |
Binding ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
47 |
0.39 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z50417-3-O |
Leishmania Infantum (cluster #3 Of 4), Other |
Other |
7000 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
7900 |
0.27 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.28 |
ADME/T ≤ 10μM |
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
4760 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
6.7 |
-52.4 |
3 |
3 |
1 |
50 |
379.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.93 |
17.63 |
-43.36 |
2 |
2 |
1 |
25 |
529.959 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
8.93 |
17.63 |
-43.45 |
2 |
2 |
1 |
25 |
529.959 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.93 |
18.13 |
-44.7 |
2 |
2 |
1 |
25 |
529.959 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
8.93 |
18.13 |
-44.48 |
2 |
2 |
1 |
25 |
529.959 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
8.56 |
-80.64 |
4 |
4 |
2 |
51 |
357.885 |
6 |
↓
|
|
|
|
Analogs
-
1530861
-
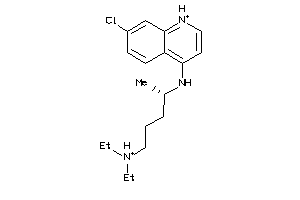
-
5850156
-
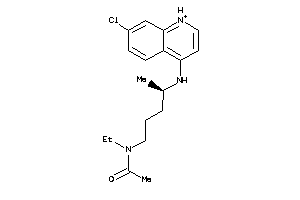
-
5850183
-
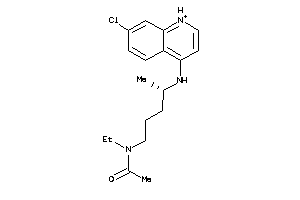
-
6424763
-
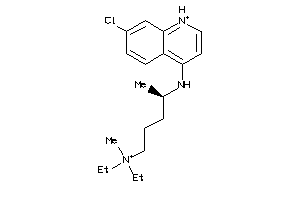
-
19144226
-
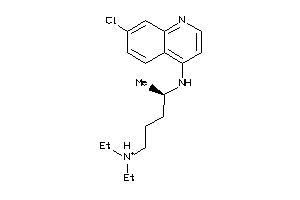
Draw
Identity
99%
90%
80%
70%
Vendors
And 66 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.59 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
-
5850156
-

-
5850183
-

-
6424763
-

-
19144226
-

-
19144231
-
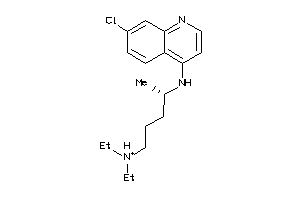
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.48 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAH-1-B |
Histone Deacetylase-like Amidohydrolase (cluster #1 Of 2), Bacterial |
Bacteria |
1300 |
0.43 |
Binding ≤ 10μM
|
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.53 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
921 |
0.44 |
Binding ≤ 10μM |
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
820 |
0.45 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
65 |
0.53 |
Binding ≤ 10μM
|
|
NCOR2-1-E |
Nuclear Receptor Corepressor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.55 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
Q7K6A1-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.52 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.53 |
Binding ≤ 10μM
|
|
Q9ZTP8-1-E |
Histone Deacetylase HD1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.55 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
Q94F81-2-E |
Histone Deacetylase HD2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
170 |
0.50 |
Functional ≤ 10μM
|
|
Z103205-2-O |
A431 (cluster #2 Of 4), Other |
Other |
4200 |
0.40 |
Functional ≤ 10μM
|
|
Z103348-2-O |
NCI-H2122 (cluster #2 Of 2), Other |
Other |
7500 |
0.38 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
820 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
|
Z80054-4-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
8100 |
0.38 |
Functional ≤ 10μM
|
|
Z80064-7-O |
CCRF-CEM (T-cell Leukemia) (cluster #7 Of 9), Other |
Other |
950 |
0.44 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2120 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-4-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #4 Of 12), Other |
Other |
1200 |
0.44 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
2400 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
750 |
0.45 |
Functional ≤ 10μM
|
|
Z80224-6-O |
MCF7 (Breast Carcinoma Cells) (cluster #6 Of 14), Other |
Other |
8500 |
0.37 |
Functional ≤ 10μM
|
|
Z80244-1-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #1 Of 7), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
6500 |
0.38 |
Functional ≤ 10μM
|
|
Z80418-9-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #9 Of 9), Other |
Other |
760 |
0.45 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
2100 |
0.42 |
Functional ≤ 10μM
|
|
Z80493-3-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
2500 |
0.41 |
Functional ≤ 10μM
|
|
Z80513-1-O |
SQ20B (cluster #1 Of 1), Other |
Other |
3000 |
0.41 |
Functional ≤ 10μM
|
|
Z80514-1-O |
St-4 (Stomach Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5200 |
0.39 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
868 |
0.45 |
Functional ≤ 10μM
|
|
Z80838-1-O |
EOL1 (Eosinophilic Cells) (cluster #1 Of 1), Other |
Other |
1000 |
0.44 |
Functional ≤ 10μM
|
|
Z80857-1-O |
Friend Leukemia Cell Line (cluster #1 Of 2), Other |
Other |
990 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-3-O |
CEM (T-cell Leukemia) (cluster #3 Of 7), Other |
Other |
1900 |
0.42 |
Functional ≤ 10μM
|
|
Z80877-1-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #1 Of 2), Other |
Other |
7240 |
0.38 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2060 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
900 |
0.45 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
9300 |
0.37 |
Functional ≤ 10μM
|
|
Z81024-2-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #2 Of 8), Other |
Other |
3448 |
0.40 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
2600 |
0.41 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-3-O |
LNCaP (Prostate Carcinoma) (cluster #3 Of 5), Other |
Other |
580 |
0.46 |
Functional ≤ 10μM
|
|
Z81184-5-O |
LOX IMVI (Melanoma Cells) (cluster #5 Of 5), Other |
Other |
1300 |
0.43 |
Functional ≤ 10μM
|
|
Z81245-4-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #4 Of 6), Other |
Other |
1900 |
0.42 |
Functional ≤ 10μM
|
|
Z81247-6-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #6 Of 9), Other |
Other |
460 |
0.47 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
2200 |
0.42 |
Functional ≤ 10μM
|
|
Z81280-3-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
3480 |
0.40 |
Functional ≤ 10μM
|
|
Z81281-4-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #4 Of 5), Other |
Other |
383 |
0.47 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
832 |
0.45 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
600 |
0.46 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3900 |
0.40 |
Functional ≤ 10μM
|
|
Z80957-2-O |
HMEC (Microvascular Endothelial Cells) (cluster #2 Of 2), Other |
Other |
1300 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
4.41 |
-18.87 |
3 |
5 |
0 |
78 |
264.325 |
8 |
↓
|
|