|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
4030 |
0.84 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
4030 |
0.84 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
-0.54 |
-5.48 |
2 |
2 |
0 |
40 |
124.139 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.87 |
2.41 |
-47.04 |
0 |
2 |
-1 |
40 |
123.131 |
0 |
↓
|
|
|
|
Analogs
-
4482687
-
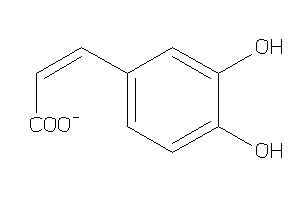
Draw
Identity
99%
90%
80%
70%
Vendors
And 63 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8710 |
0.54 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
7870 |
0.55 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.59 |
Binding ≤ 10μM
|
|
O49150-1-E |
5-lipoxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.58 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3060 |
0.59 |
Binding ≤ 10μM
|
|
Z100766-1-O |
Radical Scavenging Activity (cluster #1 Of 2), Other |
Other |
2900 |
0.60 |
Functional ≤ 10μM
|
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
190 |
0.72 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
1.43 |
-49.59 |
2 |
4 |
-1 |
81 |
179.151 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
1640 |
0.74 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
1640 |
0.74 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
-1.11 |
-8.57 |
2 |
3 |
0 |
57 |
152.149 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.65 |
-4.1 |
-53.04 |
3 |
5 |
-1 |
100 |
195.15 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
7410 |
0.80 |
Binding ≤ 10μM |
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
540 |
0.97 |
Binding ≤ 10μM |
|
CAH6-5-E |
Carbonic Anhydrase VI (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
520 |
0.98 |
Binding ≤ 10μM |
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4390 |
0.83 |
Binding ≤ 10μM |
|
Q9BEG3-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.84 |
Functional ≤ 10μM
|
|
LUXP-1-B |
Autoinducer 2-binding Periplasmic Protein LuxP (cluster #1 Of 2), Bacterial |
Bacteria |
2000 |
0.89 |
Functional ≤ 10μM
|
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
490 |
0.98 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
990 |
0.93 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
5660 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.73 |
-2.62 |
-8.63 |
3 |
3 |
0 |
61 |
126.111 |
0 |
↓
|
|
|
|
Analogs
-
31910660
-
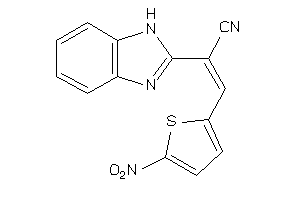
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103204-3-O |
A427 (cluster #3 Of 4), Other |
Other |
150 |
0.45 |
Functional ≤ 10μM
|
|
Z50185-3-O |
Staphylococcus Aureus (cluster #3 Of 4), Other |
Other |
6400 |
0.35 |
Functional ≤ 10μM
|
|
Z50186-2-O |
Staphylococcus Epidermidis (cluster #2 Of 2), Other |
Other |
3200 |
0.37 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
190 |
0.45 |
Functional ≤ 10μM
|
|
Z80112-1-O |
DAN-G (cluster #1 Of 2), Other |
Other |
600 |
0.41 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
260 |
0.44 |
Functional ≤ 10μM
|
|
Z80435-1-O |
RT-112 (cluster #1 Of 1), Other |
Other |
360 |
0.43 |
Functional ≤ 10μM
|
|
Z80436-1-O |
RT-4 (cluster #1 Of 1), Other |
Other |
270 |
0.44 |
Functional ≤ 10μM
|
|
Z80473-1-O |
SISO (cluster #1 Of 2), Other |
Other |
480 |
0.42 |
Functional ≤ 10μM
|
|
Z80546-1-O |
TERT-RPE1 (Retinal Pigmented Epithelial Cells) (cluster #1 Of 1), Other |
Other |
390 |
0.43 |
Functional ≤ 10μM
|
|
Z80604-1-O |
YAPC (cluster #1 Of 2), Other |
Other |
210 |
0.45 |
Functional ≤ 10μM
|
|
Z81126-1-O |
KYSE-150 Cell Line (cluster #1 Of 1), Other |
Other |
330 |
0.43 |
Functional ≤ 10μM
|
|
Z81128-1-O |
KYSE-520 Cell Line (cluster #1 Of 1), Other |
Other |
110 |
0.46 |
Functional ≤ 10μM
|
|
Z81130-1-O |
KYSE-70 Cell Line (cluster #1 Of 1), Other |
Other |
160 |
0.45 |
Functional ≤ 10μM
|
|
Z81146-1-O |
LCLC-103H Cell Line (cluster #1 Of 1), Other |
Other |
160 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
0.16 |
-17.06 |
1 |
6 |
0 |
98 |
296.311 |
3 |
↓
|
|