|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
-2.06 |
-15.43 |
1 |
5 |
0 |
63 |
309.398 |
3 |
↓
|
|
|
|
Analogs
-
4473722
-
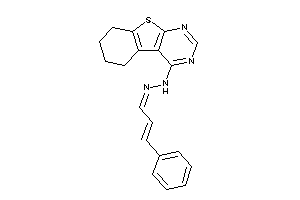
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
1400 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.64 |
-0.84 |
-11.78 |
1 |
4 |
0 |
50 |
334.448 |
4 |
↓
|
|
|
|
Analogs
-
33908130
-
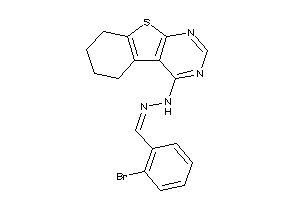
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.65 |
-1.61 |
-11.73 |
1 |
4 |
0 |
50 |
387.306 |
3 |
↓
|
|
|
|
Analogs
-
5054820
-
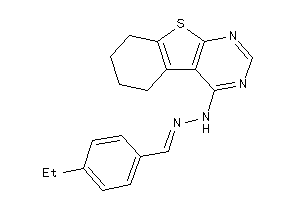
-
33924061
-
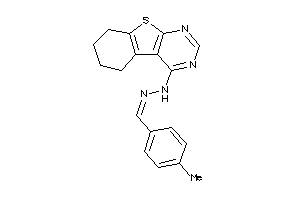
-
38194854
-
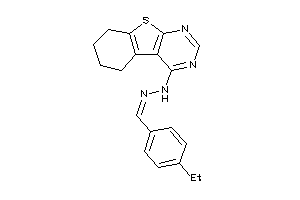
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
-0.23 |
-10.61 |
1 |
4 |
0 |
50 |
350.491 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50422-1-O |
Entamoeba Histolytica (cluster #1 Of 5), Other |
Other |
4270 |
0.40 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8970 |
0.37 |
Functional ≤ 10μM |
|
Z50473-1-O |
Plasmodium Berghei (cluster #1 Of 5), Other |
Other |
4070 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.78 |
8.49 |
-18.62 |
1 |
7 |
0 |
93 |
260.253 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
0.96 |
5.63 |
-19.78 |
1 |
7 |
0 |
96 |
260.253 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.78 |
8.96 |
-41.98 |
2 |
7 |
1 |
94 |
261.261 |
5 |
↓
|
|
|
|
Analogs
-
7997571
-
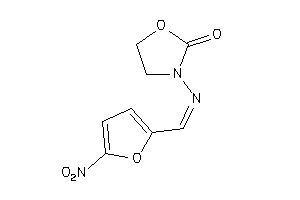
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
400 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
2.48 |
-32.24 |
0 |
8 |
0 |
101 |
225.16 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
-1.68 |
-9.33 |
3 |
5 |
0 |
76 |
334.354 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
-1.42 |
-81.02 |
5 |
5 |
2 |
79 |
336.37 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
-1.57 |
-38.28 |
4 |
5 |
1 |
78 |
335.362 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
1000 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
7.21 |
-9.17 |
0 |
4 |
0 |
59 |
200.197 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.00 |
7.42 |
-36.8 |
1 |
4 |
1 |
60 |
201.205 |
2 |
↓
|
|
|
|
Analogs
-
4073970
-
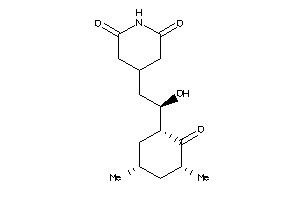
-
4536598
-
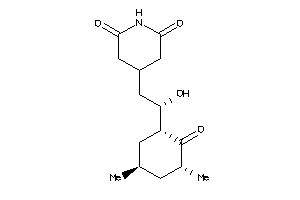
-
8579261
-
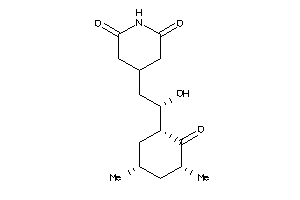
-
11592912
-
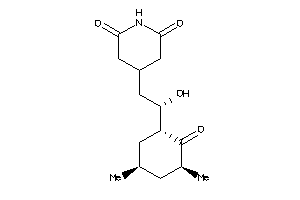
-
11592913
-
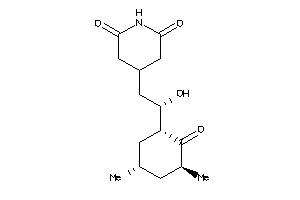
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
Z50347-2-O |
Saccharomyces Cerevisiae (cluster #2 Of 3), Other |
Other |
88 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
400 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z80026-2-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
3600 |
0.38 |
Functional ≤ 10μM |
|
Z80224-6-O |
MCF7 (Breast Carcinoma Cells) (cluster #6 Of 14), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3500 |
0.38 |
Functional ≤ 10μM
|
|
Z80485-2-O |
SK-MEL-28 (Melanoma Cells) (cluster #2 Of 6), Other |
Other |
1000 |
0.42 |
Functional ≤ 10μM
|
|
Z80490-1-O |
SK-MES-1 (cluster #1 Of 4), Other |
Other |
2700 |
0.39 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 7), Other |
Other |
530 |
0.44 |
Functional ≤ 10μM
|
|
Z80653-1-O |
9L (Glioma Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z80874-4-O |
CEM (T-cell Leukemia) (cluster #4 Of 7), Other |
Other |
80 |
0.50 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
2500 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-7-O |
Jurkat (Acute Leukemic T-cells) (cluster #7 Of 10), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-3-O |
LNCaP (Prostate Carcinoma) (cluster #3 Of 5), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM
|
|
Z81247-6-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #6 Of 9), Other |
Other |
533 |
0.44 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM |
|
Z80005-2-O |
3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
912 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
3.22 |
-17 |
2 |
5 |
0 |
83 |
281.352 |
3 |
↓
|
|
|
|
Analogs
-
4245662
-
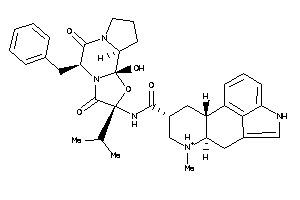
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
-5.46 |
-64.32 |
4 |
10 |
1 |
119 |
612.751 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
2900 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
14.94 |
-32.01 |
1 |
10 |
-1 |
144 |
581.649 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
5.72 |
14.85 |
-31 |
1 |
10 |
-1 |
144 |
581.649 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.72 |
17.46 |
-45.92 |
2 |
10 |
0 |
145 |
582.657 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
2900 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.72 |
14.61 |
-35.56 |
1 |
10 |
-1 |
144 |
581.649 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.72 |
17.13 |
-44.76 |
2 |
10 |
0 |
145 |
582.657 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.72 |
17.14 |
-42.97 |
2 |
10 |
0 |
145 |
582.657 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-3-O |
Trypanosoma Cruzi (cluster #3 Of 8), Other |
Other |
2300 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
6.26 |
-9.86 |
1 |
4 |
0 |
54 |
245.307 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.91 |
5.14 |
-41.82 |
0 |
4 |
-1 |
56 |
244.299 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
3700 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
-3.04 |
-11.92 |
3 |
3 |
0 |
50 |
207.302 |
5 |
↓
|
|
|
|
Analogs
-
12959788
-
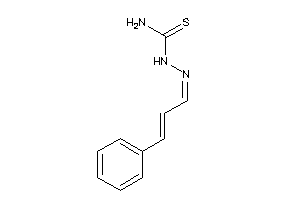
-
12959792
-
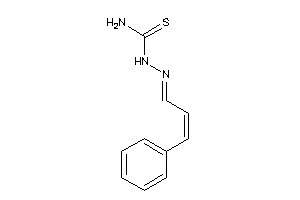
-
8579322
-
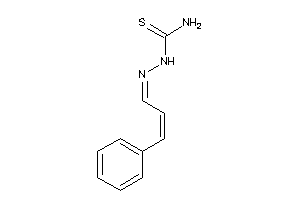
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
7400 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.11 |
5.62 |
-11.77 |
3 |
3 |
0 |
50 |
205.286 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.21 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
400 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
-1.13 |
-43.11 |
1 |
8 |
1 |
66 |
533.48 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q53I59-1-E |
Cyclic Nucleotide Specific Phosphodiesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
3300 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
4.39 |
-17.5 |
0 |
5 |
0 |
67 |
330.39 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
-1.18 |
-20.69 |
1 |
12 |
0 |
115 |
700.791 |
12 |
↓
|
|
|
|
Analogs
-
531272
-
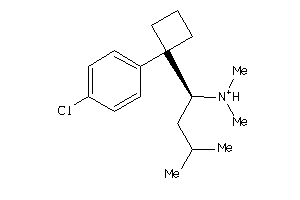
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
10.53 |
-34.64 |
1 |
1 |
1 |
4 |
280.863 |
5 |
↓
|
|
|
|
Analogs
-
4759
-
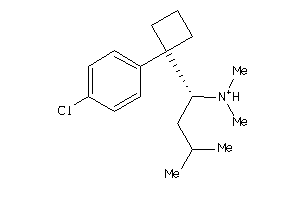
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.00 |
10.74 |
-33.26 |
1 |
1 |
1 |
4 |
280.863 |
5 |
↓
|
|
|
|
Analogs
-
22064264
-
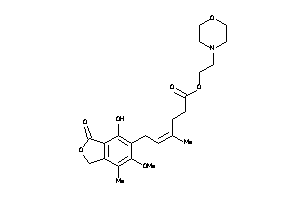
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
7000 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
6.63 |
-15.37 |
1 |
8 |
0 |
95 |
433.501 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.82 |
7.41 |
-55.75 |
0 |
8 |
-1 |
97 |
432.493 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
2000 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
7.95 |
-49.42 |
3 |
2 |
1 |
29 |
310.248 |
7 |
↓
|
|
|
|
Analogs
-
38735350
-
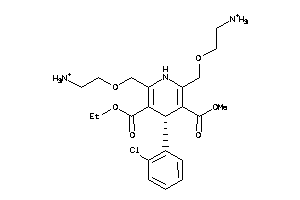
-
38735353
-
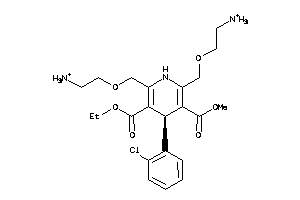
Draw
Identity
99%
90%
80%
70%
Vendors
And 67 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2344 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7943 |
0.26 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5370 |
0.26 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z101865-1-O |
Leishmania Chagasi (cluster #1 Of 1), Other |
Other |
5350 |
0.26 |
Functional ≤ 10μM
|
|
Z50457-1-O |
Leishmania Amazonensis (cluster #1 Of 3), Other |
Other |
2930 |
0.28 |
Functional ≤ 10μM
|
|
Z50458-3-O |
Leishmania Braziliensis (cluster #3 Of 4), Other |
Other |
3130 |
0.28 |
Functional ≤ 10μM
|
|
Z50460-3-O |
Leishmania Major (cluster #3 Of 4), Other |
Other |
3350 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8070 |
0.25 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
7.61 |
-58.02 |
4 |
7 |
1 |
102 |
409.89 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.83 |
6.31 |
-39.45 |
2 |
7 |
-1 |
106 |
407.874 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
7.16 |
-13.89 |
3 |
7 |
0 |
100 |
408.882 |
10 |
↓
|
|
|
|
Analogs
-
38735350
-

-
38735353
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 60 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2344 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7943 |
0.26 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5370 |
0.26 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z101865-1-O |
Leishmania Chagasi (cluster #1 Of 1), Other |
Other |
5350 |
0.26 |
Functional ≤ 10μM
|
|
Z50457-1-O |
Leishmania Amazonensis (cluster #1 Of 3), Other |
Other |
2930 |
0.28 |
Functional ≤ 10μM
|
|
Z50458-3-O |
Leishmania Braziliensis (cluster #3 Of 4), Other |
Other |
3130 |
0.28 |
Functional ≤ 10μM
|
|
Z50460-3-O |
Leishmania Major (cluster #3 Of 4), Other |
Other |
3350 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8070 |
0.25 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
7.61 |
-60.61 |
4 |
7 |
1 |
102 |
409.89 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
7.17 |
-11.79 |
3 |
7 |
0 |
100 |
408.882 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.83 |
6.31 |
-39.43 |
2 |
7 |
-1 |
106 |
407.874 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KD4DL-1-E |
Lysine-specific Demethylase 4D-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3550 |
0.42 |
Binding ≤ 10μM |
|
Z101879-1-O |
Leptomonas Seymouri (cluster #1 Of 1), Other |
Other |
400 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.40 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
210 |
0.52 |
Functional ≤ 10μM
|
|
Z80002-2-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
800 |
0.47 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
7 |
0.63 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
1650 |
0.45 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
9960 |
0.39 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
1130 |
0.46 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
4210 |
0.42 |
Functional ≤ 10μM
|
|
Z80980-1-O |
HOS (Osteosarcoma Cells) (cluster #1 Of 2), Other |
Other |
1400 |
0.46 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
1800 |
0.45 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
250 |
0.51 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 2), Other |
Other |
2100 |
0.44 |
ADME/T ≤ 10μM
|
|
Z80156-4-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #4 Of 4), Other |
Other |
270 |
0.51 |
ADME/T ≤ 10μM
|
|
Z80901-1-O |
HaCaT (Keratinocytes) (cluster #1 Of 2), Other |
Other |
700 |
0.48 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
8.03 |
-10.63 |
0 |
3 |
0 |
43 |
242.274 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
2700 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
2.47 |
-22.89 |
3 |
7 |
0 |
109 |
214.206 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q53I59-1-E |
Cyclic Nucleotide Specific Phosphodiesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
4800 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
-3.8 |
-9.27 |
2 |
5 |
0 |
66 |
332.454 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q53I59-1-E |
Cyclic Nucleotide Specific Phosphodiesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.30 |
Binding ≤ 10μM
|
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
6400 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
9.34 |
-10.58 |
1 |
7 |
0 |
94 |
355.423 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.45 |
9.08 |
-46.01 |
0 |
7 |
-1 |
92 |
354.415 |
6 |
↓
|
|
|
|
Analogs
-
3978828
-
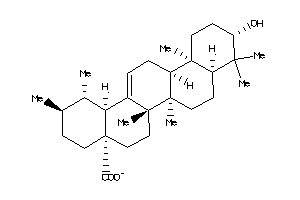
-
3978829
-
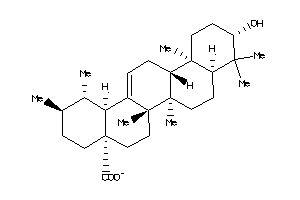
-
4273370
-
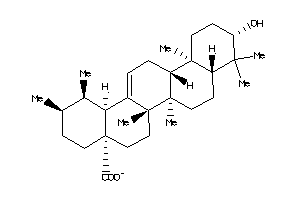
-
4273371
-
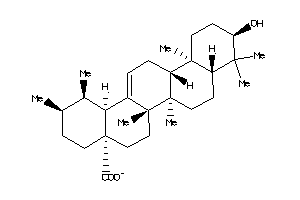
-
4273372
-
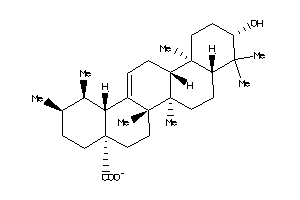
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
DPOLB-2-E |
DNA Polymerase Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.22 |
Binding ≤ 10μM
|
|
PA21B-3-E |
Phospholipase A2 Group 1B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.23 |
Binding ≤ 10μM
|
|
PA2A-1-E |
Phospholipase A2 Isozyme PLA-A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GA-3-E |
Phospholipase A2, Membrane Associated (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GD-2-E |
Group IID Secretory Phospholipase A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GE-2-E |
Group IIE Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GF-2-E |
Group IIF Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.23 |
Binding ≤ 10μM
|
|
PTN2-2-E |
T-cell Protein-tyrosine Phosphatase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6700 |
0.22 |
Binding ≤ 10μM |
|
PYGM-1-E |
Muscle Glycogen Phosphorylase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.21 |
Binding ≤ 10μM |
|
Q7T3S7-1-E |
Phospholipase A2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1430 |
0.25 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-4-O |
Trypanosoma Brucei (cluster #4 Of 6), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50420-3-O |
Trypanosoma Brucei Brucei (cluster #3 Of 7), Other |
Other |
2200 |
0.24 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50472-2-O |
Toxoplasma Gondii (cluster #2 Of 4), Other |
Other |
1000 |
0.25 |
Functional ≤ 10μM
|
|
Z50607-8-O |
Human Immunodeficiency Virus 1 (cluster #8 Of 10), Other |
Other |
1800 |
0.24 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
4400 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
12.05 |
-49.78 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.79 |
10.29 |
-5.45 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
340 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
1300 |
0.46 |
Functional ≤ 10μM
|
|
Z80120-4-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #4 Of 5), Other |
Other |
1440 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-11-O |
A549 (Lung Carcinoma Cells) (cluster #11 Of 11), Other |
Other |
550 |
0.49 |
Functional ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 4), Other |
Other |
8100 |
0.40 |
Functional ≤ 10μM
|
|
Z81225-2-O |
M109 (Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1000 |
0.47 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
9650 |
0.39 |
Functional ≤ 10μM
|
|
Z100094-1-O |
MLF (Lung Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
4900 |
0.41 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
8.58 |
-23.57 |
1 |
2 |
1 |
20 |
233.294 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
6700 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
7.43 |
-8.22 |
1 |
2 |
0 |
29 |
252.704 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.70 |
7.83 |
-23.51 |
2 |
2 |
1 |
30 |
253.712 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
10.29 |
-18.57 |
1 |
8 |
0 |
114 |
357.347 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
3500 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
3.95 |
-28.54 |
3 |
7 |
0 |
109 |
240.244 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
700 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
0.84 |
-17.67 |
0 |
6 |
0 |
63 |
354.358 |
4 |
↓
|
|
|
|
Analogs
-
7997796
-
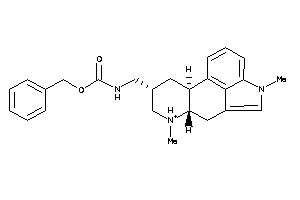
-
537987
-
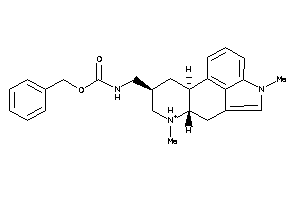
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.38 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.35 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.36 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.36 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.38 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.24 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
4000 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
12.27 |
-49.04 |
2 |
5 |
1 |
48 |
404.534 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-8-O |
Trypanosoma Cruzi (cluster #8 Of 8), Other |
Other |
9000 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
3.3 |
-19.01 |
0 |
4 |
0 |
58 |
178.191 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.31 |
19.49 |
-13.94 |
3 |
5 |
0 |
69 |
563.73 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
8.31 |
20 |
-48.44 |
4 |
5 |
1 |
70 |
564.738 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
4.29 |
-16.48 |
2 |
9 |
0 |
138 |
252.186 |
4 |
↓
|
|
|
|
Analogs
-
1758871
-
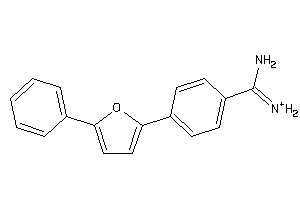
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
Z50420-4-O |
Trypanosoma Brucei Brucei (cluster #4 Of 7), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z50466-7-O |
Trypanosoma Cruzi (cluster #7 Of 8), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z50468-4-O |
Giardia Intestinalis (cluster #4 Of 4), Other |
Other |
440 |
0.39 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z50725-1-O |
Trypanosoma Brucei Rhodesiense (cluster #1 Of 7), Other |
Other |
4500 |
0.33 |
Functional ≤ 10μM
|
|
Z80035-5-O |
B16 (Melanoma Cells) (cluster #5 Of 7), Other |
Other |
9200 |
0.31 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
6400 |
0.32 |
Functional ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 6), Other |
Other |
6700 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
5.7 |
-76.45 |
8 |
5 |
2 |
116 |
306.369 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8430 |
0.16 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.87 |
20.97 |
-50.94 |
2 |
9 |
1 |
115 |
612.747 |
14 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.87 |
19.19 |
-18.9 |
1 |
9 |
0 |
114 |
611.739 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
8430 |
0.16 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.87 |
21.02 |
-52.19 |
2 |
9 |
1 |
115 |
612.747 |
14 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.87 |
19.22 |
-17.68 |
1 |
9 |
0 |
114 |
611.739 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
8.65 |
-8.33 |
0 |
3 |
0 |
43 |
250.253 |
2 |
↓
|
|
|
|
Analogs
-
5855933
-
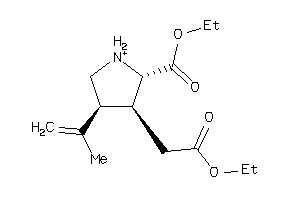
-
34599078
-
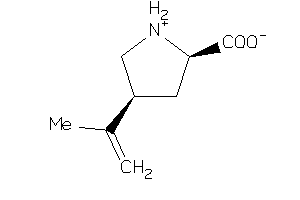
-
1481864
-
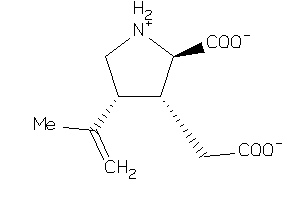
-
2008786
-
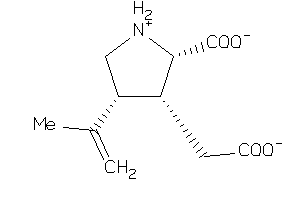
-
3872987
-
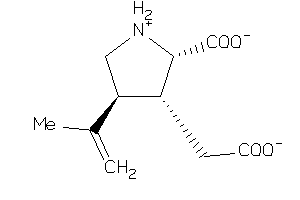
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
77 |
0.66 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.48 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.49 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.49 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1714 |
0.54 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
177 |
0.63 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
53 |
0.68 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.75 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7510 |
0.48 |
Functional ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.57 |
Functional ≤ 10μM
|
|
Z50597-4-O |
Rattus Norvegicus (cluster #4 Of 5), Other |
Other |
18 |
0.72 |
Binding ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
3000 |
0.52 |
Functional ≤ 10μM
|
|
Z50597-6-O |
Rattus Norvegicus (cluster #6 Of 12), Other |
Other |
3700 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.81 |
5.21 |
-88.22 |
2 |
5 |
-1 |
97 |
212.225 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.29 |
Binding ≤ 10μM
|
|
PGTB1-1-E |
Geranylgeranyl Transferase Type I Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
Q5EI73-1-E |
Protein Farnesyltransferase Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM
|
|
Q5EI74-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.32 |
Binding ≤ 10μM
|
|
FNTA-1-E |
Protein Farnesyltransferase/geranylgeranyltransferase Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.29 |
Functional ≤ 10μM
|
|
FNTB-1-E |
Protein Farnesyltransferase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.29 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4060 |
0.22 |
ADME/T ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
4 |
0.35 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
2 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
13.62 |
-14.28 |
2 |
5 |
0 |
66 |
489.406 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.09 |
14.08 |
-48.31 |
3 |
5 |
1 |
67 |
490.414 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.09 |
14.29 |
-131.87 |
4 |
5 |
2 |
69 |
491.422 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
37121385
-
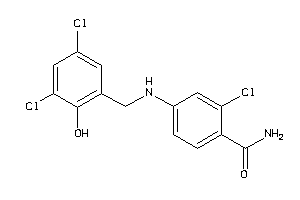
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
8000 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.65 |
-1.34 |
-10.05 |
2 |
3 |
0 |
49 |
351.016 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.29 |
Binding ≤ 10μM
|
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.29 |
Binding ≤ 10μM
|
|
CACB1-1-E |
Voltage-dependent L-type Calcium Channel Subunit Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.29 |
Binding ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.31 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.31 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
12.82 |
-48.51 |
1 |
5 |
1 |
35 |
469.552 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.97 |
10.52 |
-9.52 |
0 |
5 |
0 |
34 |
468.544 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.97 |
12.37 |
-49.34 |
1 |
5 |
1 |
35 |
469.552 |
8 |
↓
|
|
|
|
Analogs
-
4521067
-
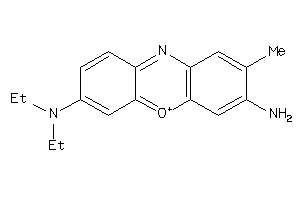
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
11.31 |
-9.05 |
0 |
4 |
0 |
31 |
324.448 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
0.96 |
10.23 |
-19.87 |
0 |
4 |
1 |
32 |
324.448 |
5 |
↓
|
|
|
|
Analogs
-
1530638
-
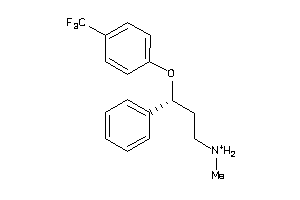
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
130 |
0.44 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM |
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5820 |
0.33 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
176 |
0.43 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
563 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.51 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.32 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
605 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.35 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.34 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
33 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
9.16 |
-43.81 |
2 |
2 |
1 |
26 |
310.339 |
7 |
↓
|
|