|
|
Analogs
-
1530611
-
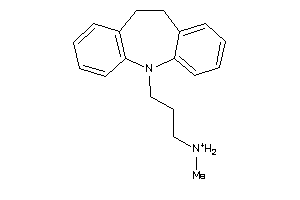
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
94 |
0.47 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
160 |
0.45 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT5B-1-E |
Serotonin 5b (5-HT5b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.49 |
Binding ≤ 10μM
|
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.43 |
Binding ≤ 10μM
|
|
ADA1A-2-E |
Alpha-1a Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.49 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
46 |
0.49 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.49 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
ADCY1-1-E |
Brain Adenylate Cyclase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.45 |
Binding ≤ 10μM
|
|
ADCY2-2-E |
Adenylate Cyclase Type II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.45 |
Binding ≤ 10μM
|
|
ADCY8-1-E |
Adenylate Cyclase Type VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.45 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.49 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.49 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.49 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
645 |
0.41 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
645 |
0.41 |
Binding ≤ 10μM
|
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
645 |
0.41 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
645 |
0.41 |
Binding ≤ 10μM
|
|
HRH1-2-E |
Histamine H1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.50 |
Binding ≤ 10μM
|
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.50 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.50 |
Binding ≤ 10μM
|
|
HRH4-1-E |
Histamine H4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.50 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5470 |
0.35 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.57 |
Binding ≤ 10μM |
|
SC6A2-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
74 |
0.48 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
42 |
0.49 |
Binding ≤ 10μM
|
|
SC6A4-3-E |
Serotonin Transporter (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.36 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.36 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.36 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.36 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.36 |
Functional ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.48 |
Functional ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.41 |
Functional ≤ 10μM
|
|
DRD2-14-E |
Dopamine D2 Receptor (cluster #14 Of 24), Eukaryotic |
Eukaryotes |
645 |
0.41 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
65 |
0.48 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
46 |
0.49 |
Binding ≤ 10μM |
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
12 |
0.53 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1259 |
0.39 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
300 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
85 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
5000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
12.58 |
-35.72 |
1 |
2 |
1 |
8 |
281.423 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 101 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.31 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
610 |
0.36 |
ADME/T ≤ 10μM
|
|
CP2DI-1-E |
Cytochrome P450 2D18 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.34 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
94 |
0.41 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
652 |
0.36 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
81 |
0.41 |
Functional ≤ 10μM
|
|
Z50460-2-O |
Leishmania Major (cluster #2 Of 4), Other |
Other |
9530 |
0.29 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
7.18 |
-40.05 |
2 |
4 |
1 |
47 |
325.432 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
7.62 |
-89.58 |
3 |
4 |
2 |
48 |
326.44 |
4 |
↓
|
|
|
|
Analogs
-
34588958
-
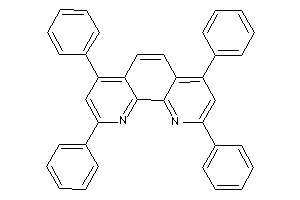
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
80 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.83 |
0.94 |
-11.71 |
0 |
2 |
0 |
25 |
360.46 |
2 |
↓
|
|
|
|
Analogs
-
34937021
-
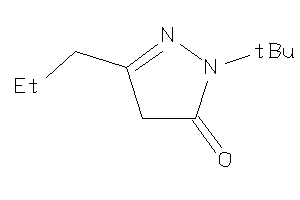
-
41655078
-
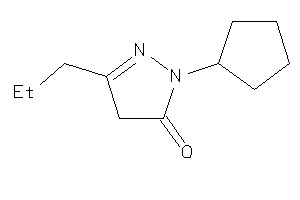
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-2-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #2 Of 3), Other |
Other |
13 |
0.85 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
5.22 |
-8.43 |
0 |
3 |
0 |
33 |
180.251 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.42 |
3 |
-5.82 |
1 |
3 |
0 |
38 |
180.251 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-2-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #2 Of 3), Other |
Other |
40 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
8.69 |
-15.45 |
1 |
3 |
0 |
38 |
250.301 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.51 |
6.69 |
-7.89 |
1 |
3 |
0 |
38 |
250.301 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-2-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #2 Of 3), Other |
Other |
2000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
5.83 |
-42.59 |
1 |
5 |
-1 |
73 |
278.291 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-2-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #2 Of 3), Other |
Other |
6 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
0.51 |
-17.17 |
1 |
4 |
0 |
54 |
278.311 |
3 |
↓
|
|
|
|
Analogs
-
38004135
-
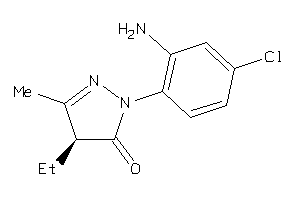
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-2-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #2 Of 3), Other |
Other |
1 |
0.90 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
5.47 |
-13.14 |
1 |
3 |
0 |
38 |
208.648 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
2.46 |
5.72 |
-9.41 |
0 |
3 |
0 |
33 |
208.648 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.46 |
3.55 |
-6.07 |
1 |
3 |
0 |
38 |
208.648 |
1 |
↓
|
|
|
|
Analogs
-
4228177
-
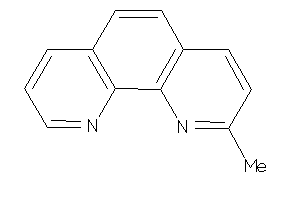
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
80 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
6.6 |
-13.89 |
0 |
2 |
0 |
26 |
208.264 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.01 |
7.07 |
-26.75 |
1 |
2 |
1 |
27 |
209.272 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-2-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #2 Of 3), Other |
Other |
3 |
0.57 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
2.2 |
-14.06 |
0 |
6 |
0 |
78 |
281.271 |
3 |
↓
|
|
|
|
Analogs
-
19721273
-
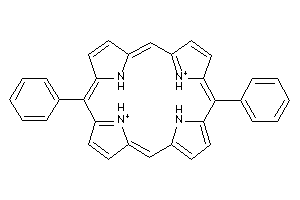
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
10 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
10.76 |
0.9 |
-13.88 |
2 |
4 |
0 |
57 |
614.752 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-6.37 |
24.54 |
-432.11 |
4 |
8 |
6 |
75 |
680.86 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
4500 |
0.27 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
33 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z80578-1-O |
UV4 (cluster #1 Of 1), Other |
Other |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z80667-1-O |
AA8 (cluster #1 Of 2), Other |
Other |
1700 |
0.29 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
800 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.56 |
11.66 |
-76.98 |
3 |
4 |
2 |
40 |
401.982 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
4500 |
0.27 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
33 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
8 |
0.40 |
Functional ≤ 10μM
|
|
Z80578-1-O |
UV4 (cluster #1 Of 1), Other |
Other |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z80667-1-O |
AA8 (cluster #1 Of 2), Other |
Other |
1700 |
0.29 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
800 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.56 |
11.53 |
-77.73 |
3 |
4 |
2 |
40 |
401.982 |
9 |
↓
|
|
|
|
Analogs
-
1530861
-
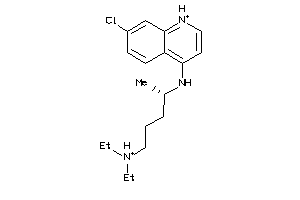
-
5850156
-
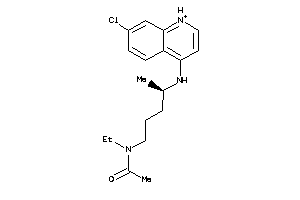
-
5850183
-
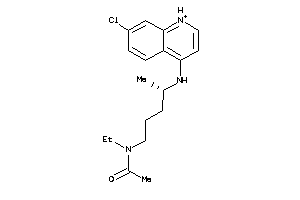
-
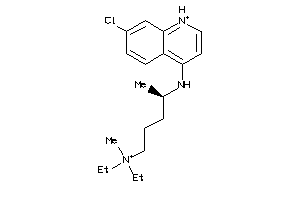
6424763
-
-
19144226
-
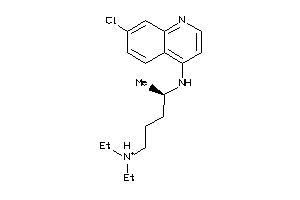
Draw
Identity
99%
90%
80%
70%
Vendors
And 66 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.59 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
-
5850156
-

-
5850183
-

-
6424763
-

-
19144226
-

-
19144231
-
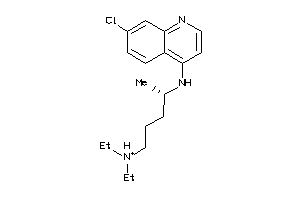
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.36 |
Binding ≤ 10μM
|
|
NQO2-1-E |
Quinone Reductase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Binding ≤ 10μM
|
|
HRP1-1-E |
Histidine-rich Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
400 |
0.41 |
Binding ≤ 10μM
|
|
Z100275-1-O |
Hemozoin (cluster #1 Of 1), Other |
Other |
77 |
0.45 |
Functional ≤ 10μM
|
|
Z100498-2-O |
Hepatocytes (cluster #2 Of 2), Other |
Other |
1900 |
0.36 |
Functional ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
1700 |
0.37 |
Functional ≤ 10μM
|
|
Z102013-2-O |
Plasmodium Malariae (cluster #2 Of 2), Other |
Other |
14 |
0.50 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
167 |
0.43 |
Functional ≤ 10μM
|
|
Z50424-2-O |
Cryptosporidium Parvum (cluster #2 Of 2), Other |
Other |
27 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z50468-2-O |
Giardia Intestinalis (cluster #2 Of 4), Other |
Other |
401 |
0.41 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
72 |
0.45 |
Functional ≤ 10μM
|
|
Z50474-1-O |
Plasmodium Yoelii (cluster #1 Of 1), Other |
Other |
106 |
0.44 |
Functional ≤ 10μM
|
|
Z80136-1-O |
FM3A (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
9670 |
0.32 |
Functional ≤ 10μM
|
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.34 |
ADME/T ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
32 |
0.48 |
ADME/T ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
64 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.32 |
ADME/T ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
600 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
7700 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
10.55 |
-81.48 |
3 |
3 |
2 |
31 |
321.896 |
8 |
↓
|
|
|
|
Analogs
-
1280206
-
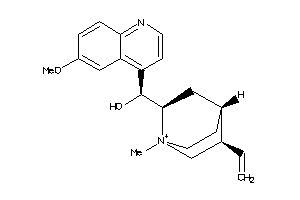
-
1319234
-
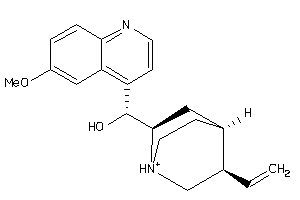
-
4289113
-
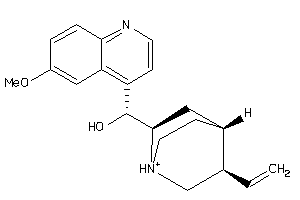
-
4521307
-
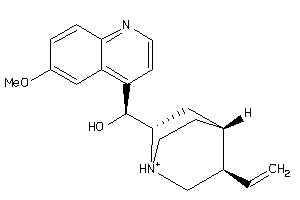
-
6484901
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.30 |
Binding ≤ 10μM
|
|
KCND2-1-E |
Potassium Voltage-gated Channel Subfamily D Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.30 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.31 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.30 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5600 |
0.31 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.32 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
4 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
51 |
0.43 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
5000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
7.17 |
-40.03 |
2 |
4 |
1 |
47 |
325.432 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
7.61 |
-90.06 |
3 |
4 |
2 |
48 |
326.44 |
4 |
↓
|
|
|
|
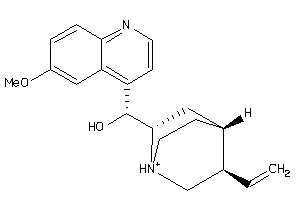
Analogs
-
4289113
-

-
4521307
-

-
6484901
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.30 |
Binding ≤ 10μM
|
|
KCND2-1-E |
Potassium Voltage-gated Channel Subfamily D Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.30 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.31 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.30 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5600 |
0.31 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.32 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
4 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
51 |
0.43 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
5000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
6.3 |
-41.9 |
2 |
4 |
1 |
47 |
325.432 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
6.73 |
-91.82 |
3 |
4 |
2 |
48 |
326.44 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 75 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.30 |
Binding ≤ 10μM
|
|
KCND2-1-E |
Potassium Voltage-gated Channel Subfamily D Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.30 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.31 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.30 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5600 |
0.31 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.32 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
4 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
51 |
0.43 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
5000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
7.47 |
-38.76 |
2 |
4 |
1 |
47 |
325.432 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
7.91 |
-89.66 |
3 |
4 |
2 |
48 |
326.44 |
4 |
↓
|
|
|
|
Analogs
-
402416
-
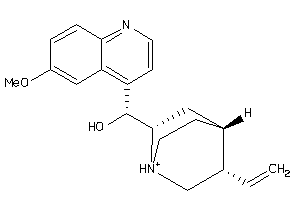
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7300 |
0.30 |
Binding ≤ 10μM
|
|
KCND2-1-E |
Potassium Voltage-gated Channel Subfamily D Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.33 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.30 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.31 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.30 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.32 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5600 |
0.31 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.32 |
ADME/T ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
4 |
0.49 |
Functional ≤ 10μM
|
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
51 |
0.43 |
Functional ≤ 10μM
|
|
Z81138-1-O |
ScN2a (Scrapie-infected Neuroblastoma Cells) (cluster #1 Of 3), Other |
Other |
5000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
6.29 |
-42.11 |
2 |
4 |
1 |
47 |
325.432 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
6.72 |
-92.39 |
3 |
4 |
2 |
48 |
326.44 |
4 |
↓
|
|