|
|
Analogs
-
967970
-
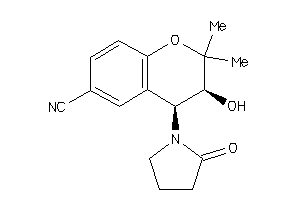
-
967971
-
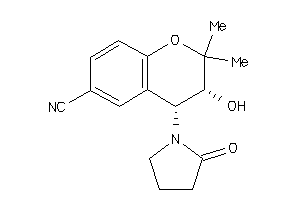
-
1224
-
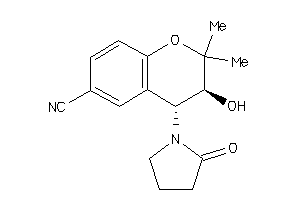
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
98 |
0.47 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
600 |
0.41 |
Functional ≤ 10μM |
|
Z50588-2-O |
Canis Familiaris (cluster #2 Of 7), Other |
Other |
1960 |
0.38 |
Functional ≤ 10μM |
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
420 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
74 |
0.48 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
5.1 |
-12.95 |
1 |
5 |
0 |
74 |
286.331 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8200 |
0.24 |
Binding ≤ 10μM
|
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.25 |
Binding ≤ 10μM
|
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.24 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
800 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
5.42 |
-26.25 |
3 |
8 |
0 |
106 |
471.988 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.43 |
5.82 |
-69.29 |
2 |
8 |
-1 |
106 |
470.98 |
9 |
↓
|
|
|
|
Analogs
-
3995739
-
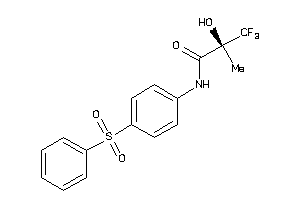
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
Z50512-6-O |
Cavia Porcellus (cluster #6 Of 7), Other |
Other |
470 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
2.76 |
-14.78 |
2 |
5 |
0 |
83 |
373.352 |
5 |
↓
|
|
|
|
Analogs
-
3995738
-
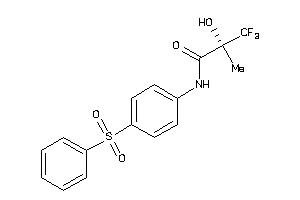
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Binding ≤ 10μM |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.35 |
Functional ≤ 10μM
|
|
Z50512-6-O |
Cavia Porcellus (cluster #6 Of 7), Other |
Other |
470 |
0.35 |
Functional ≤ 10μM
|
|
Z50512-6-O |
Cavia Porcellus (cluster #6 Of 7), Other |
Other |
1000 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
2.76 |
-14.69 |
2 |
5 |
0 |
83 |
373.352 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-2-B |
Beta-lactamase (cluster #2 Of 6), Bacterial |
Bacteria |
49 |
0.31 |
Binding ≤ 10μM
|
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
CTRA-3-E |
Alpha-chymotrypsin (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
KCND3-1-E |
Voltage-gated Potassium Channel Subunit Kv4.3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
MDHM-1-E |
Malate Dehydrogenase, Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
8 |
0.34 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2 |
0.37 |
Functional ≤ 10μM
|
|
Z81047-1-O |
Human T-cell Line (cluster #1 Of 2), Other |
Other |
1000 |
0.25 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
5.51 |
-67.24 |
2 |
8 |
-1 |
120 |
493.005 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
4.40 |
4.95 |
-23.65 |
3 |
8 |
0 |
117 |
494.013 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.77 |
7.45 |
-21.59 |
3 |
8 |
0 |
114 |
494.013 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Binding ≤ 10μM
|
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Binding ≤ 10μM
|
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Binding ≤ 10μM
|
|
KCND3-1-E |
Voltage-gated Potassium Channel Subunit Kv4.3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Binding ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7447 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
1.45 |
-65.23 |
2 |
9 |
-1 |
136 |
444.537 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
1.94 |
0.9 |
-22.02 |
3 |
9 |
0 |
134 |
445.545 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.31 |
3.39 |
-20.04 |
3 |
9 |
0 |
130 |
445.545 |
7 |
↓
|
|