|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.20 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4060 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.60 |
19.21 |
-66.32 |
0 |
6 |
-1 |
76 |
513.621 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.60 |
19.62 |
-59.12 |
1 |
6 |
0 |
77 |
514.629 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.60 |
19.88 |
-93.84 |
2 |
6 |
1 |
78 |
515.637 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.23 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Binding ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
12.6 |
-119.08 |
0 |
8 |
-2 |
113 |
433.512 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.08 |
13.62 |
-64.47 |
1 |
8 |
-1 |
115 |
434.52 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
1.37 |
-104.59 |
0 |
9 |
-2 |
121 |
427.468 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.34 |
Binding ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM |
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM |
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM |
|
AGTR1-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Functional ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.37 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM |
|
Z80902-1-O |
HASMC (Aortic Smooth Muscle Cells) (cluster #1 Of 2), Other |
Other |
320 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.44 |
11.96 |
-56.34 |
0 |
7 |
-1 |
85 |
427.532 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.44 |
11.97 |
-14.39 |
1 |
7 |
0 |
87 |
428.54 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.30 |
Binding ≤ 10μM |
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
97 |
0.26 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
97 |
0.26 |
Binding ≤ 10μM
|
|
Q2XUH7-1-E |
Angiotensin Type II Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
34 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
18.82 |
-70.42 |
0 |
7 |
-1 |
82 |
507.614 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.36 |
Binding ≤ 10μM
|
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.38 |
Binding ≤ 10μM |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.38 |
Binding ≤ 10μM |
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.38 |
Binding ≤ 10μM |
|
AGTR1-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.37 |
Functional ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
89 |
0.33 |
Functional ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
82 |
0.33 |
Functional ≤ 10μM |
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.38 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM |
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
19 |
0.36 |
Functional ≤ 10μM
|
|
Z80902-1-O |
HASMC (Aortic Smooth Muscle Cells) (cluster #1 Of 2), Other |
Other |
1 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
9.35 |
-51.76 |
1 |
7 |
-1 |
91 |
421.912 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.87 |
9.43 |
-17.19 |
2 |
7 |
0 |
93 |
422.92 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
420 |
0.26 |
Binding ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.28 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.28 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
1.02 |
-75.76 |
1 |
8 |
-1 |
113 |
482.944 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.32 |
Binding ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.30 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.29 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.29 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.86 |
11.89 |
-20.71 |
1 |
8 |
0 |
107 |
582.792 |
12 |
↓
|
|
Ref
Reference (pH 7)
|
6.86 |
12.51 |
-74.12 |
0 |
8 |
-1 |
110 |
581.784 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.28 |
Binding ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.28 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.28 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
15.66 |
-69.48 |
2 |
7 |
-1 |
104 |
479.56 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.36 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.36 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.32 |
2.66 |
-42.82 |
0 |
7 |
-1 |
87 |
419.896 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Functional ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
11.37 |
-54.09 |
0 |
8 |
-1 |
96 |
415.525 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.53 |
11.63 |
-54.43 |
1 |
8 |
0 |
98 |
416.533 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.32 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.32 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.01 |
4.03 |
-56.79 |
0 |
9 |
-1 |
113 |
553.668 |
11 |
↓
|
|
|
|
Analogs
-
38422268
-
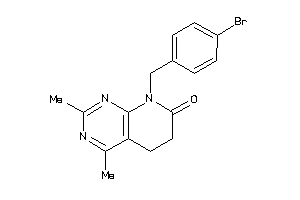
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.34 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
11.11 |
-51.78 |
0 |
8 |
-1 |
99 |
410.461 |
4 |
↓
|
|
|
|
Analogs
-
12503190
-
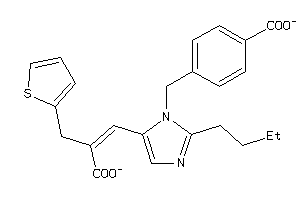
-
29319828
-
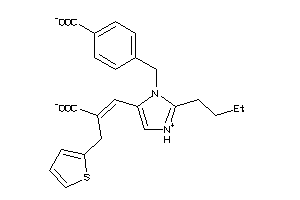
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
12.07 |
-108.21 |
1 |
6 |
-1 |
99 |
409.487 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.33 |
0.54 |
-105.97 |
1 |
6 |
-1 |
95 |
409.487 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
391 |
0.37 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
391 |
0.37 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
391 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
9.77 |
-112.71 |
0 |
6 |
-2 |
98 |
326.352 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.25 |
10.21 |
-109.38 |
1 |
6 |
-1 |
99 |
327.36 |
8 |
↓
|
|
|
|
Analogs
-
1552601
-
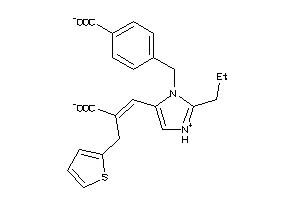
-
12503190
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.38 |
Binding ≤ 10μM |
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
363 |
0.30 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.38 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
13.17 |
-110.32 |
1 |
6 |
-1 |
99 |
423.514 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.89 |
12.91 |
-114.55 |
0 |
6 |
-2 |
98 |
422.506 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.33 |
Binding ≤ 10μM |
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.33 |
Binding ≤ 10μM |
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.33 |
Binding ≤ 10μM
|
|
AGTR1-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
13.26 |
-58.4 |
0 |
10 |
-1 |
113 |
509.594 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.16 |
13.43 |
-43.11 |
2 |
10 |
1 |
116 |
511.61 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.16 |
13.53 |
-42.49 |
2 |
10 |
1 |
116 |
511.61 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGTR1-1-E |
Type-1 Angiotensin II Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
AGTRA-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Binding ≤ 10μM
|
|
AGTR1-1-E |
Angiotensin II Type 1a (AT-1a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Functional ≤ 10μM
|
|
AGTR2-1-E |
Angiotensin II Type 2 (AT-2) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.34 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
26 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
11.47 |
-95.77 |
0 |
8 |
-2 |
111 |
434.887 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.10 |
11.86 |
-71.42 |
1 |
8 |
-1 |
112 |
435.895 |
8 |
↓
|
|