|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7100 |
0.60 |
Binding ≤ 10μM
|
|
AMPM2-2-E |
Methionine Aminopeptidase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
3.8 |
-6.08 |
1 |
3 |
0 |
42 |
159.192 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.66 |
-0.38 |
-97.46 |
1 |
7 |
-2 |
126 |
258.189 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.66 |
-0.71 |
-92.61 |
1 |
7 |
-2 |
126 |
258.189 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
-0.24 |
-34.9 |
1 |
8 |
-1 |
132 |
260.185 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.69 |
0.32 |
-37.72 |
1 |
8 |
-1 |
132 |
260.185 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.23 |
2.19 |
-11.07 |
2 |
8 |
0 |
129 |
261.193 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
-0.81 |
-44.22 |
1 |
6 |
-1 |
89 |
258.257 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.85 |
-0.68 |
-43.42 |
1 |
6 |
-1 |
89 |
258.257 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.40 |
1.9 |
-8.41 |
2 |
6 |
0 |
86 |
259.265 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
0.36 |
-44.74 |
1 |
5 |
-1 |
86 |
241.226 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.51 |
0.33 |
-44.86 |
1 |
5 |
-1 |
86 |
241.226 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.51 |
0.32 |
-45.36 |
1 |
5 |
-1 |
86 |
241.226 |
2 |
↓
|
|
|
|
Analogs
-
33849788
-
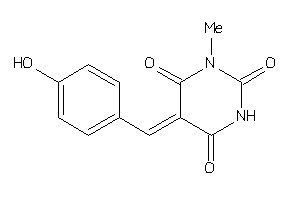
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.81 |
-1.86 |
-41.05 |
1 |
6 |
-1 |
95 |
245.214 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.81 |
-1.77 |
-42.06 |
1 |
6 |
-1 |
95 |
245.214 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.35 |
0.82 |
-8.64 |
2 |
6 |
0 |
92 |
246.222 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
2.5 |
-36.49 |
1 |
4 |
-1 |
69 |
249.246 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.26 |
2.55 |
-38.34 |
1 |
4 |
-1 |
69 |
249.246 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.80 |
4.22 |
-9.56 |
2 |
4 |
0 |
66 |
250.254 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
1.56 |
-8.42 |
2 |
5 |
0 |
83 |
234.186 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.27 |
-3.99 |
-41.54 |
2 |
6 |
-1 |
106 |
231.187 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.27 |
-3.22 |
-89.45 |
1 |
6 |
-2 |
109 |
230.179 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.19 |
-1.33 |
-9.53 |
3 |
6 |
0 |
103 |
232.195 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
-0.46 |
-38.08 |
1 |
8 |
-1 |
132 |
260.185 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.25 |
2.1 |
-13.47 |
2 |
8 |
0 |
129 |
261.193 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.71 |
-0.37 |
-38.74 |
1 |
8 |
-1 |
132 |
260.185 |
2 |
↓
|
|
|
|
Analogs
-
24258146
-
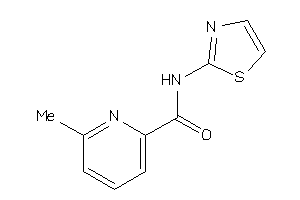
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM1-2-E |
Methionine Aminopeptidase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8100 |
0.51 |
Binding ≤ 10μM |
|
CTR4-1-E |
Cationic Amino Acid Transporter 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.52 |
Binding ≤ 10μM
|
|
AMPM-2-B |
Methionine Aminopeptidase (cluster #2 Of 3), Bacterial |
Bacteria |
5000 |
0.53 |
Binding ≤ 10μM
|
|
AMPM1-1-F |
Methionine Aminopeptidase 1 (cluster #1 Of 1), Fungal |
Fungi |
7000 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
3.35 |
-12.95 |
1 |
4 |
0 |
55 |
205.242 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.78 |
1.79 |
-52 |
0 |
4 |
-1 |
61 |
204.234 |
2 |
↓
|
|