|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2400 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
10.01 |
-58.93 |
2 |
11 |
-1 |
161 |
472.437 |
6 |
↓
|
|
|
|
Analogs
-
541750
-
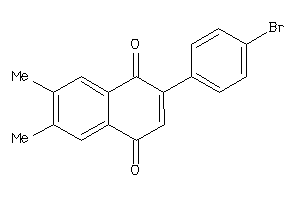
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9330 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
1.81 |
-7.15 |
0 |
2 |
0 |
34 |
234.254 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8220 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
5.89 |
-8.62 |
1 |
3 |
0 |
54 |
250.253 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.91 |
6.67 |
-48.32 |
0 |
3 |
-1 |
57 |
249.245 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
7.65 |
-60.87 |
5 |
9 |
1 |
132 |
579.743 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
49 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
9.11 |
-66.57 |
5 |
8 |
1 |
119 |
506.668 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.03 |
8.87 |
-18.14 |
4 |
8 |
0 |
117 |
505.66 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
5930 |
0.16 |
Binding ≤ 10μM
|
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
108 |
0.21 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1440 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.40 |
11.07 |
-40.78 |
2 |
8 |
1 |
89 |
633.898 |
22 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.86 |
14.06 |
-11.18 |
2 |
8 |
0 |
83 |
632.89 |
24 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.19 |
Binding ≤ 10μM
|
|
BACE2-1-E |
Beta Secretase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.19 |
Binding ≤ 10μM
|
|
PEN2-1-E |
Gamma-secretase Subunit PEN-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.46 |
10.74 |
-21.78 |
6 |
10 |
0 |
160 |
672.867 |
19 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
21 |
0.24 |
Binding ≤ 10μM
|
|
BACE2-1-E |
Beta Secretase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
10.11 |
-73.73 |
4 |
8 |
1 |
105 |
608.706 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
605 |
0.38 |
Binding ≤ 10μM
|
|
CATD-1-E |
Cathepsin D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
11.83 |
-36.3 |
3 |
4 |
1 |
60 |
320.501 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.79 |
11.88 |
-37.57 |
3 |
4 |
1 |
58 |
320.501 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.79 |
11.93 |
-5.49 |
2 |
4 |
0 |
56 |
319.493 |
5 |
↓
|
|