|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
7800 |
0.89 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
90 |
1.23 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8200 |
0.89 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5930 |
0.91 |
Binding ≤ 10μM |
|
TYRO-3-F |
Tyrosinase (cluster #3 Of 8), Fungal |
Fungi |
9810 |
0.88 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
-1.23 |
-4.89 |
2 |
2 |
0 |
40 |
110.112 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2370 |
0.79 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
3530 |
0.76 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
600 |
0.87 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
3170 |
0.77 |
Binding ≤ 10μM |
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4530 |
0.75 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
1.26 |
-50.95 |
1 |
3 |
-1 |
60 |
137.114 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
9200 |
0.78 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
108 |
1.08 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6700 |
0.80 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
2.04 |
-6.13 |
1 |
2 |
0 |
44 |
119.123 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 52 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
550 |
0.80 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
3670 |
0.69 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
510 |
0.80 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4410 |
0.68 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2130 |
0.72 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
-1.59 |
-50.1 |
2 |
4 |
-1 |
81 |
153.113 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 85 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
9200 |
1.01 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
5500 |
1.05 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2700 |
1.11 |
Binding ≤ 10μM
|
|
CAH4-8-E |
Carbonic Anhydrase IV (cluster #8 Of 16), Eukaryotic |
Eukaryotes |
9500 |
1.00 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
8800 |
1.01 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
1.63 |
-3.7 |
1 |
1 |
0 |
20 |
94.113 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
8360 |
0.71 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
8630 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
3.69 |
-6.18 |
0 |
2 |
0 |
18 |
138.166 |
2 |
↓
|
|
|
|
Analogs
-
1684
-
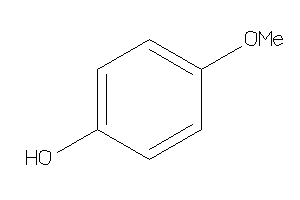
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
8210 |
0.71 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
4020 |
0.76 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
3350 |
0.77 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4150 |
0.75 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
3.07 |
-4.42 |
0 |
2 |
0 |
18 |
138.166 |
2 |
↓
|
|
|
|
Analogs
-
1727807
-
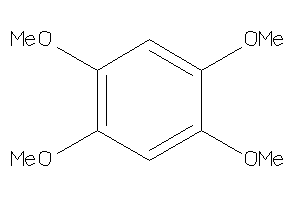
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
5960 |
0.61 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
7800 |
0.60 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
1430 |
0.68 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4060 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
2.98 |
-6.9 |
0 |
3 |
0 |
28 |
168.192 |
3 |
↓
|
|
|
|
Analogs
-
391883
-
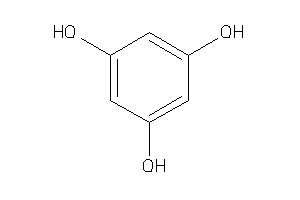
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
7500 |
0.90 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
7700 |
0.89 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
8700 |
0.89 |
Binding ≤ 10μM
|
|
CAH5B-8-E |
Carbonic Anhydrase VB (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
7100 |
0.90 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3600 |
0.95 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6420 |
0.91 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.96 |
-1.22 |
-5.57 |
2 |
2 |
0 |
40 |
110.112 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.32 |
1.74 |
-46.85 |
0 |
2 |
-1 |
40 |
109.104 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 48 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
-0.44 |
-6.18 |
2 |
2 |
0 |
40 |
110.112 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 113 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
9920 |
0.70 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
8800 |
0.71 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7200 |
0.72 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
7120 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
3.11 |
-51.26 |
1 |
3 |
-1 |
60 |
137.114 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 77 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
7610 |
0.72 |
Binding ≤ 10μM
|
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
9800 |
0.70 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
6270 |
0.73 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7790 |
0.72 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
6160 |
0.73 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9320 |
0.70 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
870 |
0.85 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
7280 |
0.72 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6610 |
0.73 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
7780 |
0.72 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
9200 |
0.71 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
7130 |
0.72 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5700 |
0.73 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4080 |
0.75 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
3730 |
0.76 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8460 |
0.71 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
2.22 |
-47.43 |
1 |
3 |
-1 |
60 |
137.114 |
1 |
↓
|
|