|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 58 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM
|
|
CAH12-4-E |
Carbonic Anhydrase XII (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4100 |
0.69 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9230 |
0.64 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
6200 |
0.66 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7100 |
0.66 |
Binding ≤ 10μM
|
|
CAH7-4-E |
Carbonic Anhydrase VII (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9100 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
0.54 |
-10.06 |
2 |
3 |
0 |
49 |
151.165 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 75 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.88 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.53 |
-0.49 |
-4.34 |
3 |
2 |
0 |
46 |
109.128 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 52 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-4-E |
Carbonic Anhydrase XII (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
380 |
0.64 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
620 |
0.62 |
Binding ≤ 10μM
|
|
CAH14-3-E |
Carbonic Anhydrase XIV (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
590 |
0.62 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
570 |
0.62 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
420 |
0.64 |
Binding ≤ 10μM
|
|
CAH4-4-E |
Carbonic Anhydrase IV (cluster #4 Of 16), Eukaryotic |
Eukaryotes |
58 |
0.72 |
Binding ≤ 10μM
|
|
CAH5A-5-E |
Carbonic Anhydrase VA (cluster #5 Of 10), Eukaryotic |
Eukaryotes |
48 |
0.73 |
Binding ≤ 10μM
|
|
CAH5B-6-E |
Carbonic Anhydrase VB (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
61 |
0.72 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
640 |
0.62 |
Binding ≤ 10μM
|
|
CAH7-4-E |
Carbonic Anhydrase VII (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
360 |
0.64 |
Binding ≤ 10μM
|
|
CAH9-5-E |
Carbonic Anhydrase IX (cluster #5 Of 11), Eukaryotic |
Eukaryotes |
510 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
3.84 |
-49.82 |
4 |
2 |
1 |
40 |
187.266 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 85 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
9200 |
1.01 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
5500 |
1.05 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2700 |
1.11 |
Binding ≤ 10μM
|
|
CAH4-8-E |
Carbonic Anhydrase IV (cluster #8 Of 16), Eukaryotic |
Eukaryotes |
9500 |
1.00 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
8800 |
1.01 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
1.63 |
-3.7 |
1 |
1 |
0 |
20 |
94.113 |
0 |
↓
|
|
|
|
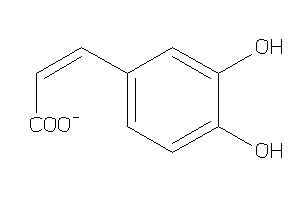
Analogs
-
4482687
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 63 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8710 |
0.54 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
7870 |
0.55 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.59 |
Binding ≤ 10μM
|
|
O49150-1-E |
5-lipoxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.58 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3060 |
0.59 |
Binding ≤ 10μM
|
|
Z100766-1-O |
Radical Scavenging Activity (cluster #1 Of 2), Other |
Other |
2900 |
0.60 |
Functional ≤ 10μM
|
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
190 |
0.72 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
1.43 |
-49.59 |
2 |
4 |
-1 |
81 |
179.151 |
2 |
↓
|
|
|
|
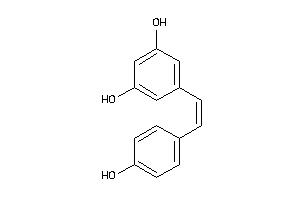
Analogs
-
12353732
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AHR-1-E |
Aryl Hydrocarbon Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
169 |
0.56 |
Binding ≤ 10μM
|
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2210 |
0.47 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
950 |
0.50 |
Binding ≤ 10μM
|
|
CAH13-7-E |
Carbonic Anhydrase XIII (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
4090 |
0.44 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9360 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
2770 |
0.46 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9090 |
0.42 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4470 |
0.44 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
4750 |
0.44 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4640 |
0.44 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8070 |
0.42 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4350 |
0.44 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
810 |
0.50 |
Binding ≤ 10μM
|
|
ESR1-5-E |
Estrogen Receptor Alpha (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
785 |
0.50 |
Binding ≤ 10μM
|
|
NQO2-4-E |
Quinone Reductase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.50 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3490 |
0.45 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2840 |
0.46 |
Functional ≤ 10μM
|
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
6918 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
5000 |
0.44 |
Functional ≤ 10μM
|
|
Z80224-12-O |
MCF7 (Breast Carcinoma Cells) (cluster #12 Of 14), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 9), Other |
Other |
4000 |
0.44 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
0.79 |
-8.46 |
3 |
3 |
0 |
61 |
228.247 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
7800 |
0.89 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
90 |
1.23 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8200 |
0.89 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5930 |
0.91 |
Binding ≤ 10μM |
|
TYRO-3-F |
Tyrosinase (cluster #3 Of 8), Fungal |
Fungi |
9810 |
0.88 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
-1.23 |
-4.89 |
2 |
2 |
0 |
40 |
110.112 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
9200 |
0.78 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
108 |
1.08 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6700 |
0.80 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
2.04 |
-6.13 |
1 |
2 |
0 |
44 |
119.123 |
0 |
↓
|
|
|
|
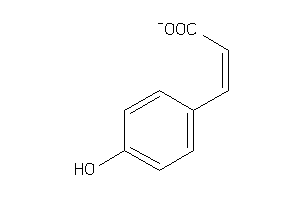
Analogs
-
4632590
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 65 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1070 |
0.70 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
8010 |
0.59 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
6680 |
0.60 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
980 |
0.70 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7570 |
0.60 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
9600 |
0.59 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
5960 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
7760 |
0.60 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6720 |
0.60 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5230 |
0.62 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
5330 |
0.62 |
Binding ≤ 10μM
|
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
190 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
2.5 |
-50.43 |
1 |
3 |
-1 |
60 |
163.152 |
2 |
↓
|
|
|
|
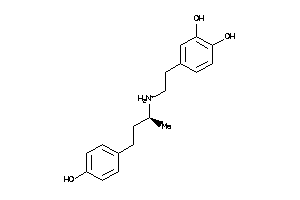
Analogs
-
57278
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
9530 |
0.32 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
4 |
0.53 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
470 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
3.38 |
-54.79 |
5 |
4 |
1 |
77 |
302.394 |
7 |
↓
|
|
|
|
Analogs
-
3911
-
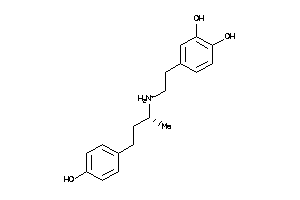
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
9530 |
0.32 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
4 |
0.53 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
470 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
3.38 |
-54.72 |
5 |
4 |
1 |
77 |
302.394 |
7 |
↓
|
|
|
|
Analogs
-
38413756
-
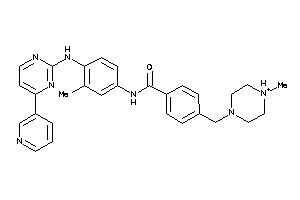
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A1Z199-1-E |
BCR/ABL P210 Fusion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
750 |
0.23 |
Binding ≤ 10μM
|
|
ABCG2-2-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4898 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.30 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.31 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
473 |
0.24 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
520 |
0.24 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
980 |
0.23 |
Binding ≤ 10μM
|
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7450 |
0.19 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
468 |
0.24 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.27 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
30 |
0.28 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
528 |
0.24 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
4553 |
0.20 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
392 |
0.24 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
109 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
76 |
0.27 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.20 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.20 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
960 |
0.23 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.28 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.30 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.19 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.21 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.21 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.21 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
97 |
0.27 |
Binding ≤ 10μM
|
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.27 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.25 |
Binding ≤ 10μM
|
|
MELK-1-E |
Maternal Embryonic Leucine Zipper Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.22 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.20 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.21 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.21 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.27 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.19 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.22 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.20 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.20 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.27 |
Functional ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.20 |
Functional ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.27 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.19 |
Functional ≤ 10μM
|
|
Z80036-1-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #1 Of 3), Other |
Other |
9100 |
0.19 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
60 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
6 |
0.31 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
10000 |
0.19 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
13.15 |
-49.4 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
13.03 |
-52.36 |
3 |
8 |
1 |
87 |
494.623 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.89 |
10.78 |
-14.7 |
2 |
8 |
0 |
86 |
493.615 |
7 |
↓
|
|