|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.33 |
-2.14 |
-122.57 |
6 |
6 |
2 |
91 |
396.376 |
15 |
↓
|
|
|
|
Analogs
-
1532734
-
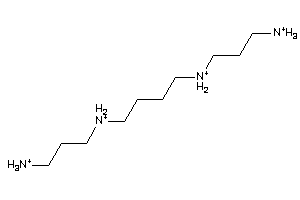
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 12), Eukaryotic |
Eukaryotes |
1400 |
0.82 |
Binding ≤ 10μM
|
|
CAH14-2-E |
Carbonic Anhydrase XIV (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.84 |
Binding ≤ 10μM
|
|
CAH15-3-E |
Carbonic Anhydrase 15 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.70 |
Binding ≤ 10μM
|
|
CAH2-4-E |
Carbonic Anhydrase II (cluster #4 Of 15), Eukaryotic |
Eukaryotes |
1110 |
0.83 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
112 |
0.97 |
Binding ≤ 10μM
|
|
CAH5A-4-E |
Carbonic Anhydrase VA (cluster #4 Of 10), Eukaryotic |
Eukaryotes |
1220 |
0.83 |
Binding ≤ 10μM
|
|
CAH5B-5-E |
Carbonic Anhydrase VB (cluster #5 Of 9), Eukaryotic |
Eukaryotes |
1440 |
0.82 |
Binding ≤ 10μM
|
|
CAH6-3-E |
Carbonic Anhydrase VI (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
1410 |
0.82 |
Binding ≤ 10μM
|
|
CAH7-3-E |
Carbonic Anhydrase VII (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
1230 |
0.83 |
Binding ≤ 10μM
|
|
CAH9-4-E |
Carbonic Anhydrase IX (cluster #4 Of 11), Eukaryotic |
Eukaryotes |
1370 |
0.82 |
Binding ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
4400 |
0.75 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
113 |
0.97 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.83 |
-0.71 |
-175.27 |
8 |
3 |
3 |
72 |
148.274 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.83 |
-1.09 |
-97.55 |
7 |
3 |
2 |
70 |
147.266 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.83 |
-2.05 |
-90.25 |
7 |
3 |
2 |
67 |
147.266 |
7 |
↓
|
|
|
|
Analogs
-
1532612
-
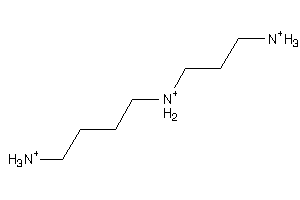
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH14-2-E |
Carbonic Anhydrase XIV (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
860 |
0.61 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
10 |
0.80 |
Binding ≤ 10μM
|
|
CAH5A-4-E |
Carbonic Anhydrase VA (cluster #4 Of 10), Eukaryotic |
Eukaryotes |
840 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B-5-E |
Carbonic Anhydrase VB (cluster #5 Of 9), Eukaryotic |
Eukaryotes |
830 |
0.61 |
Binding ≤ 10μM
|
|
CAH6-3-E |
Carbonic Anhydrase VI (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
990 |
0.60 |
Binding ≤ 10μM
|
|
CAH7-3-E |
Carbonic Anhydrase VII (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
710 |
0.61 |
Binding ≤ 10μM
|
|
CASP2-3-E |
Caspase-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.51 |
Binding ≤ 10μM
|
|
Z100250-2-O |
Calf Thymus DNA (cluster #2 Of 2), Other |
Other |
4300 |
0.54 |
Binding ≤ 10μM
|
|
Z104302-6-O |
Glutamate NMDA Receptor (cluster #6 Of 7), Other |
Other |
5200 |
0.53 |
Binding ≤ 10μM
|
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
390 |
0.64 |
Functional ≤ 10μM
|
|
Z80608-2-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #2 Of 4), Other |
Other |
200 |
0.67 |
Functional ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
920 |
0.60 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
280 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.13 |
0.14 |
-262.68 |
10 |
4 |
4 |
88 |
206.378 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-0.62 |
-97.52 |
8 |
4 |
2 |
85 |
204.362 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-1.59 |
-88.23 |
8 |
4 |
2 |
82 |
204.362 |
11 |
↓
|
|
|
|
Analogs
-
34360644
-
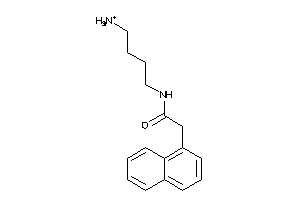
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-4-E |
Carbonic Anhydrase XII (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
190 |
0.35 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
1030 |
0.31 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
780 |
0.32 |
Binding ≤ 10μM
|
|
CAH2-4-E |
Carbonic Anhydrase II (cluster #4 Of 15), Eukaryotic |
Eukaryotes |
1130 |
0.31 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
18 |
0.40 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
1030 |
0.31 |
Binding ≤ 10μM
|
|
CAH5B-6-E |
Carbonic Anhydrase VB (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
1050 |
0.31 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
110 |
0.36 |
Binding ≤ 10μM
|
|
CAH7-4-E |
Carbonic Anhydrase VII (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
CAH9-5-E |
Carbonic Anhydrase IX (cluster #5 Of 11), Eukaryotic |
Eukaryotes |
120 |
0.36 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
690 |
0.32 |
Functional ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
690 |
0.32 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
690 |
0.32 |
Functional ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
690 |
0.32 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
178 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
-6.49 |
-193.13 |
8 |
5 |
3 |
89 |
373.565 |
14 |
↓
|
|
|
|
Analogs
-
1531059
-
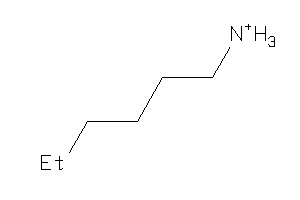
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-3-E |
Carbonic Anhydrase XII (cluster #3 Of 9), Eukaryotic |
Eukaryotes |
370 |
1.13 |
Binding ≤ 10μM
|
|
CAH13-5-E |
Carbonic Anhydrase XIII (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
690 |
1.08 |
Binding ≤ 10μM
|
|
CAH14-2-E |
Carbonic Anhydrase XIV (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
640 |
1.08 |
Binding ≤ 10μM
|
|
CAH15-3-E |
Carbonic Anhydrase 15 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
660 |
1.08 |
Binding ≤ 10μM
|
|
CAH3-3-E |
Carbonic Anhydrase III (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
600 |
1.09 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
450 |
1.11 |
Binding ≤ 10μM
|
|
CAH5A-4-E |
Carbonic Anhydrase VA (cluster #4 Of 10), Eukaryotic |
Eukaryotes |
610 |
1.09 |
Binding ≤ 10μM
|
|
CAH5B-5-E |
Carbonic Anhydrase VB (cluster #5 Of 9), Eukaryotic |
Eukaryotes |
580 |
1.09 |
Binding ≤ 10μM
|
|
CAH6-3-E |
Carbonic Anhydrase VI (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
720 |
1.07 |
Binding ≤ 10μM
|
|
CAH7-3-E |
Carbonic Anhydrase VII (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
440 |
1.11 |
Binding ≤ 10μM
|
|
CAH9-4-E |
Carbonic Anhydrase IX (cluster #4 Of 11), Eukaryotic |
Eukaryotes |
410 |
1.12 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.53 |
0.01 |
-91.02 |
6 |
2 |
2 |
55 |
118.224 |
5 |
↓
|
|
|
|
Analogs
-
29562604
-
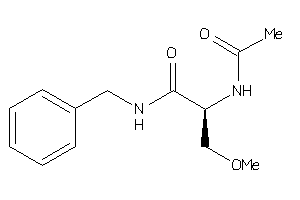
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
362 |
0.50 |
Binding ≤ 10μM
|
|
CAH12-6-E |
Carbonic Anhydrase XII (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3713 |
0.42 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1210 |
0.46 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
473 |
0.49 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
461 |
0.49 |
Binding ≤ 10μM
|
|
CAH2-14-E |
Carbonic Anhydrase II (cluster #14 Of 15), Eukaryotic |
Eukaryotes |
331 |
0.50 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
374 |
0.50 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
525 |
0.49 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
4565 |
0.42 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
341 |
0.50 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
412 |
0.50 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4446 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-7-E |
Carbonic Anhydrase IX (cluster #7 Of 11), Eukaryotic |
Eukaryotes |
353 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
-2.84 |
-9.35 |
2 |
5 |
0 |
67 |
250.298 |
6 |
↓
|
|