|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.28 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.37 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.45 |
4.1 |
-10.76 |
0 |
3 |
0 |
25 |
483.052 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.95 |
10.99 |
-8.99 |
0 |
4 |
0 |
34 |
417.384 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
CRFR2-1-E |
Corticotropin Releasing Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.25 |
17.13 |
-20.55 |
1 |
4 |
1 |
35 |
379.572 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.25 |
16.71 |
-9.3 |
0 |
4 |
0 |
34 |
378.564 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.37 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
50 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
0.69 |
-28.81 |
1 |
6 |
1 |
50 |
381.548 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
38 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
0.76 |
-9.28 |
0 |
4 |
0 |
33 |
356.901 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
31 |
0.40 |
Binding ≤ 10μM
|
|
CRFR2-1-E |
Corticotropin Releasing Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.29 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
100 |
0.38 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
70 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.90 |
14.2 |
-3.23 |
1 |
4 |
0 |
41 |
434.198 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.90 |
14.65 |
-24.97 |
2 |
4 |
1 |
42 |
435.206 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.90 |
14.35 |
-22.91 |
2 |
4 |
1 |
42 |
435.206 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
11.71 |
-9.76 |
0 |
8 |
0 |
74 |
399.495 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.43 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.02 |
2.73 |
-7.89 |
0 |
4 |
0 |
33 |
364.537 |
6 |
↓
|
|
|
|
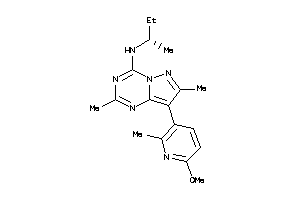
Analogs
-
40914223
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2660 |
0.31 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM |
|
NK2R-1-E |
Neurokinin 2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
129 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
9.42 |
-9 |
1 |
7 |
0 |
77 |
340.431 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2720 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
8.71 |
-4.96 |
2 |
4 |
0 |
53 |
287.75 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
3.38 |
8.1 |
-9.73 |
2 |
4 |
0 |
53 |
287.75 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.91 |
8.81 |
-30.33 |
3 |
4 |
1 |
55 |
288.758 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
310 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
9 |
-10.42 |
0 |
5 |
0 |
43 |
377.275 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
216 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
8.84 |
-10.11 |
2 |
4 |
0 |
53 |
301.777 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.31 |
9.13 |
-33.91 |
3 |
4 |
1 |
55 |
302.785 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.31 |
9.77 |
-30.3 |
3 |
4 |
1 |
55 |
302.785 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1459 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
8.73 |
-9.91 |
2 |
4 |
0 |
53 |
366.646 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
4.18 |
9.34 |
-4.55 |
2 |
4 |
0 |
53 |
366.646 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.72 |
9.64 |
-33.62 |
3 |
4 |
1 |
55 |
367.654 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.03 |
9.79 |
-7.1 |
1 |
3 |
0 |
34 |
326.484 |
6 |
↓
|
|
|
|
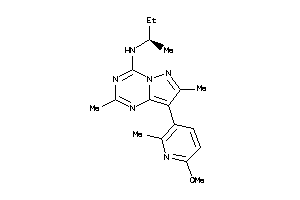
Analogs
-
34007762
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CRFR1-1-E |
Corticotropin Releasing Factor Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
9.42 |
-8.98 |
1 |
7 |
0 |
77 |
340.431 |
5 |
↓
|
|