|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
6.76 |
-39.75 |
3 |
5 |
1 |
59 |
307.603 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.23 |
4.42 |
-4.42 |
2 |
5 |
0 |
58 |
306.595 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
-2.54 |
-38.73 |
2 |
5 |
1 |
45 |
316.882 |
4 |
↓
|
|
|
|
Analogs
-
32131087
-
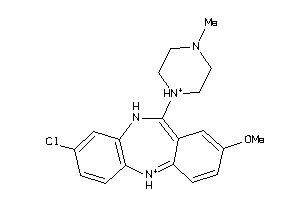
-
32131089
-
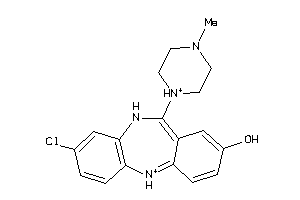
-
3995781
-
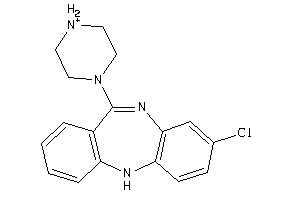
-
4292831
-
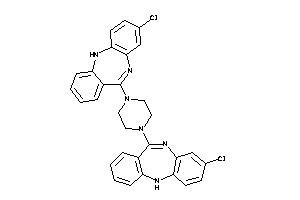
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
156 |
0.41 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.31 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.37 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH4-1-E |
Histamine H4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.44 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.32 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
390 |
0.39 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
697 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.33 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8500 |
0.31 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
631 |
0.38 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
191 |
0.41 |
Functional ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
51 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.49 |
Binding ≤ 10μM
|
|
Z102275-1-O |
Hypothalamus (cluster #1 Of 1), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.55 |
-7.08 |
1 |
4 |
0 |
35 |
326.831 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.14 |
8.88 |
-23.43 |
2 |
4 |
1 |
36 |
327.839 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.14 |
11.23 |
-89.99 |
3 |
4 |
2 |
38 |
328.847 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
23 |
0.49 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
14 |
0.50 |
Functional ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
160 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
6.5 |
-9.81 |
1 |
5 |
0 |
48 |
293.374 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.39 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.35 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.35 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
178 |
0.34 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1250 |
0.30 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2270 |
0.28 |
Binding ≤ 10μM
|
|
ADA1A-2-E |
Alpha-1a Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1320 |
0.29 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1320 |
0.29 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1320 |
0.29 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
ADA2B-2-E |
Alpha-2b Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6100 |
0.26 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.31 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
120 |
0.35 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3981 |
0.27 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
48 |
0.37 |
Binding ≤ 10μM
|
|
5HT1A-2-E |
Serotonin 1a (5-HT1a) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Functional ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
852 |
0.30 |
Binding ≤ 10μM |
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
4160 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
12.45 |
-41.93 |
1 |
7 |
1 |
71 |
386.52 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.75 |
10.24 |
-12.76 |
0 |
7 |
0 |
70 |
385.512 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
6.65 |
-36.59 |
2 |
4 |
1 |
36 |
311.384 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.06 |
8.67 |
-49.73 |
2 |
4 |
1 |
36 |
311.384 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.06 |
6.36 |
-9.06 |
1 |
4 |
0 |
35 |
310.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
181 |
0.39 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
2250 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
9.01 |
-11.25 |
1 |
6 |
0 |
61 |
320.4 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.39 |
6.8 |
-12.97 |
1 |
6 |
0 |
61 |
320.4 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.39 |
9.53 |
-110.06 |
3 |
6 |
2 |
63 |
322.416 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
7.62 |
-14.15 |
1 |
7 |
0 |
81 |
337.383 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
5.68 |
-12.46 |
1 |
4 |
0 |
35 |
310.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
6.72 |
-35.01 |
2 |
5 |
1 |
49 |
312.372 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.17 |
6.41 |
-9.13 |
1 |
5 |
0 |
48 |
311.364 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.17 |
7.69 |
-77.88 |
3 |
5 |
2 |
51 |
313.38 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.18 |
11.54 |
-11.48 |
1 |
5 |
0 |
48 |
481.427 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
42 |
0.33 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
229 |
0.30 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.31 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.34 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
32 |
0.34 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.34 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1012 |
0.27 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
316 |
0.29 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
24 |
0.34 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
476 |
0.29 |
Binding ≤ 10μM
|
|
ADRB2-2-E |
Beta-2 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
61 |
0.33 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7396 |
0.23 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
63 |
0.33 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.31 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
302 |
0.29 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6980 |
0.23 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
407 |
0.29 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6144 |
0.24 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
480 |
0.29 |
Binding ≤ 10μM
|
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
DRD2-4-E |
Dopamine D2 Receptor (cluster #4 Of 24), Eukaryotic |
Eukaryotes |
22 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
-0.53 |
-48.34 |
1 |
8 |
1 |
73 |
424.525 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2360 |
0.28 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.29 |
Binding ≤ 10μM
|
|
AA3R-4-E |
Adenosine Receptor A3 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
106 |
0.35 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.35 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.35 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
8300 |
0.25 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1585 |
0.29 |
Binding ≤ 10μM
|
|
MT3-1-E |
Metallothionein-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
S22A1-1-E |
Solute Carrier Family 22 Member 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9900 |
0.25 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
1 |
0.45 |
Binding ≤ 10μM
|
|
Z50597-5-O |
Rattus Norvegicus (cluster #5 Of 5), Other |
Other |
3000 |
0.28 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
7.64 |
-36.73 |
3 |
9 |
1 |
108 |
384.416 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.91 |
7.26 |
-15.54 |
2 |
9 |
0 |
107 |
383.408 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7100 |
0.30 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.32 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
310 |
0.38 |
Binding ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
6000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
9.51 |
-11.03 |
0 |
6 |
0 |
50 |
320.4 |
4 |
↓
|
|
|
|
Analogs
-
10519840
-
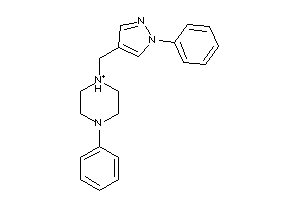
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.43 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1750 |
0.34 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1600 |
0.34 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
140 |
0.40 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
75 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
9.26 |
-8.06 |
0 |
4 |
0 |
24 |
318.424 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.42 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
38 |
0.42 |
Functional ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
64 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
9.19 |
-31.98 |
1 |
5 |
1 |
42 |
359.881 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.47 |
8.87 |
-7.76 |
0 |
5 |
0 |
41 |
358.873 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.47 |
11.42 |
-102.04 |
2 |
5 |
2 |
43 |
360.889 |
6 |
↓
|
|
|
|
Analogs
-
21835585
-
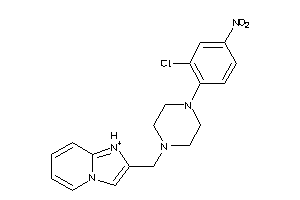
-
21403
-
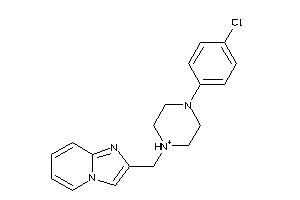
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
8.35 |
-30.47 |
1 |
4 |
1 |
25 |
327.839 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.61 |
7.94 |
-9.99 |
0 |
4 |
0 |
24 |
326.831 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6621 |
0.32 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
453 |
0.39 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.41 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.41 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1656 |
0.35 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
226 |
0.40 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.31 |
Binding ≤ 10μM
|
|
CAC1D-2-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.31 |
Binding ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.31 |
Binding ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.31 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9900 |
0.30 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
130 |
0.42 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.46 |
Functional ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
6100 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
9.91 |
-48.71 |
2 |
4 |
1 |
36 |
327.839 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.26 |
7.72 |
-8.16 |
1 |
4 |
0 |
35 |
326.831 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6600 |
0.32 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
8.45 |
-14.3 |
0 |
5 |
0 |
37 |
327.819 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.69 |
10.76 |
-58.55 |
1 |
5 |
1 |
38 |
328.827 |
3 |
↓
|
|
|
|
Analogs
-
1851395
-
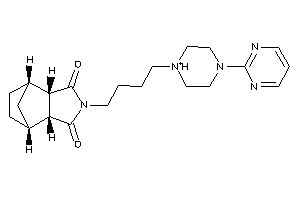
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.38 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
1260 |
0.29 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
460 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
12.37 |
-42.59 |
1 |
7 |
1 |
71 |
384.504 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.74 |
10.15 |
-11.84 |
0 |
7 |
0 |
70 |
383.496 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5400 |
0.41 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.49 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
3100 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
6.13 |
-4.96 |
1 |
3 |
0 |
22 |
241.338 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
6.65 |
-9.42 |
1 |
5 |
0 |
44 |
322.412 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.37 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8300 |
0.31 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.50 |
Binding ≤ 10μM
|
|
DRD2-23-E |
Dopamine D2 Receptor (cluster #23 Of 24), Eukaryotic |
Eukaryotes |
6300 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.34 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.35 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
66 |
0.32 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.35 |
Binding ≤ 10μM
|
|
5HT1A-2-E |
Serotonin 1a (5-HT1a) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
89 |
0.32 |
Binding ≤ 10μM
|
|
Z80094-1-O |
CHO-MG (Ovarian Cells) (cluster #1 Of 1), Other |
Other |
7 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
11.38 |
-13.41 |
0 |
6 |
0 |
49 |
422.573 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
8.79 |
-31.42 |
1 |
4 |
1 |
25 |
362.284 |
3 |
↓
|
|
|
|
Analogs
-
4628996
-
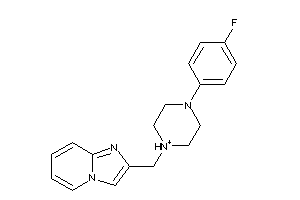
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
7.9 |
-31.24 |
1 |
4 |
1 |
25 |
311.384 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.10 |
10.21 |
-110.06 |
2 |
4 |
2 |
26 |
312.392 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.10 |
7.48 |
-10.59 |
0 |
4 |
0 |
24 |
310.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.55 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2512 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
5.75 |
-11.21 |
1 |
4 |
0 |
35 |
310.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
380 |
0.39 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2800 |
0.34 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.32 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Functional ≤ 10μM
|
|
DRD2-23-E |
Dopamine D2 Receptor (cluster #23 Of 24), Eukaryotic |
Eukaryotes |
4700 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
8.12 |
-8.75 |
0 |
4 |
0 |
24 |
326.831 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.40 |
Binding ≤ 10μM |
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.28 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.26 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.26 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.26 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
87 |
0.35 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.30 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.29 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.38 |
Binding ≤ 10μM
|
|
DRD2-8-E |
Dopamine D2 Receptor (cluster #8 Of 24), Eukaryotic |
Eukaryotes |
2900 |
0.28 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
229 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
8.86 |
-48.41 |
1 |
8 |
1 |
88 |
402.5 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.35 |
6.65 |
-16.85 |
0 |
8 |
0 |
87 |
401.492 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1530 |
0.34 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.47 |
Binding ≤ 10μM
|
|
DRD2-6-E |
Dopamine D2 Receptor (cluster #6 Of 24), Eukaryotic |
Eukaryotes |
2190 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
8.26 |
-34.11 |
2 |
5 |
1 |
49 |
320.42 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.77 |
8.3 |
-10.2 |
1 |
5 |
0 |
48 |
319.412 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.77 |
10.55 |
-39.6 |
2 |
5 |
1 |
49 |
320.42 |
4 |
↓
|
|