|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.31 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.30 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
-0.07 |
-69.14 |
0 |
8 |
-1 |
102 |
527.597 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-3-E |
Endothelin Receptor ET-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
160 |
0.29 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
4.92 |
-55.34 |
1 |
9 |
-1 |
122 |
472.543 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-3-E |
Endothelin Receptor ET-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.29 |
Binding ≤ 10μM
|
|
EDNRA-3-E |
Endothelin Receptor ET-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.29 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.25 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
6 |
-72.95 |
1 |
11 |
-1 |
148 |
550.617 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.38 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
-2.52 |
-56.15 |
0 |
8 |
-1 |
102 |
409.418 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
2.85 |
-68 |
0 |
9 |
-1 |
112 |
505.499 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
5.18 |
-61.5 |
0 |
8 |
-1 |
110 |
453.905 |
6 |
↓
|
|
|
|
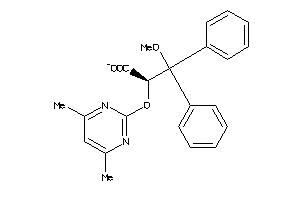
Analogs
-
3644203
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.98 |
-62.91 |
0 |
6 |
-1 |
84 |
377.42 |
7 |
↓
|
|
|
|
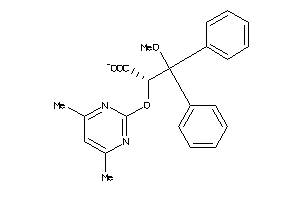
Analogs
-
538627
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
195 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.97 |
-64.38 |
0 |
6 |
-1 |
84 |
377.42 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.40 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
8.44 |
-62.47 |
0 |
6 |
-1 |
84 |
363.393 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.40 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
8.38 |
-62.22 |
0 |
6 |
-1 |
84 |
363.393 |
7 |
↓
|
|
|
|
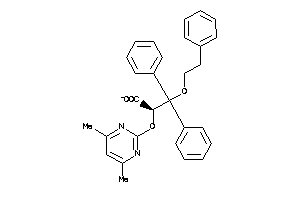
Analogs
-
13140305
-
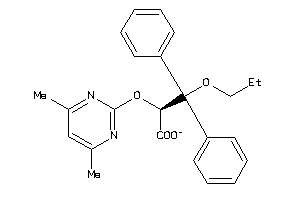
-
4675135
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.30 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
13.9 |
-67.77 |
0 |
6 |
-1 |
84 |
467.545 |
10 |
↓
|
|
|
|
Analogs
-
9660252
-
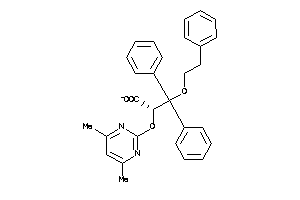
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.30 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
13.91 |
-67.98 |
0 |
6 |
-1 |
84 |
467.545 |
10 |
↓
|
|
|
|
Analogs
-
34026477
-
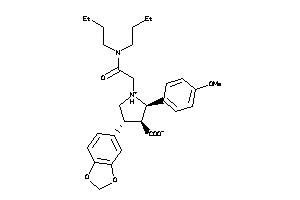
-
3812144
-
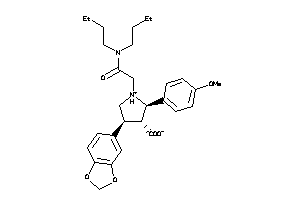
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
13.4 |
-57.6 |
1 |
8 |
0 |
93 |
510.631 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.64 |
11.89 |
-56.97 |
0 |
8 |
-1 |
91 |
509.623 |
12 |
↓
|
|
|
|
Analogs
-
34026476
-
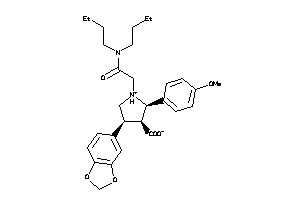
-
3812144
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
13.81 |
-58.85 |
1 |
8 |
0 |
93 |
510.631 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.64 |
12.1 |
-59.51 |
0 |
8 |
-1 |
91 |
509.623 |
12 |
↓
|
|
|
|
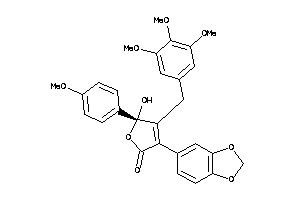
Analogs
-
13740975
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.23 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
EDNRB-1-E |
Endothelin Receptor ET-B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.24 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
8.93 |
-20.37 |
1 |
9 |
0 |
102 |
506.507 |
8 |
↓
|
|
|
|
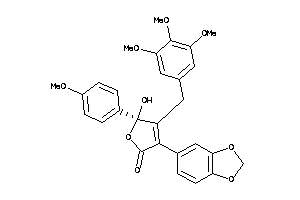
Analogs
-
13740974
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
EDNRB-1-E |
Endothelin B Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.23 |
Binding ≤ 10μM
|
|
EDNRA-1-E |
Endothelin Receptor ET-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
EDNRB-1-E |
Endothelin Receptor ET-B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.24 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
8.88 |
-20.54 |
1 |
9 |
0 |
102 |
506.507 |
8 |
↓
|
|
|
|
Analogs
-
34026476
-

-
34026477
-

Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.64 |
1.68 |
-51.98 |
1 |
8 |
0 |
92 |
510.631 |
12 |
↓
|
|