|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
756 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
-1.07 |
-6.97 |
0 |
3 |
0 |
35 |
301.143 |
2 |
↓
|
|
|
|
Analogs
-
1293163
-
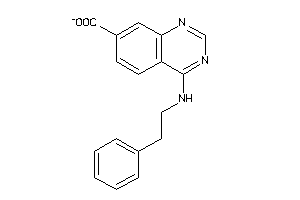
-
1293324
-
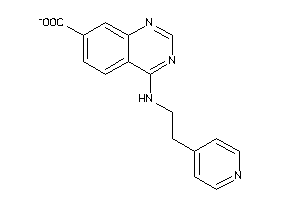
-
37862675
-
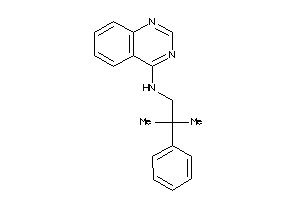
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
9.31 |
-9.32 |
1 |
3 |
0 |
38 |
249.317 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.32 |
9.78 |
-29.63 |
2 |
3 |
1 |
39 |
250.325 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
51 |
0.46 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
612 |
0.40 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
612 |
0.40 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
3375 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
1.32 |
-10.12 |
1 |
5 |
0 |
56 |
299.305 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
48 |
0.45 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
489 |
0.38 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
489 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
7.84 |
-9.82 |
1 |
5 |
0 |
56 |
313.332 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.70 |
8.31 |
-35.71 |
2 |
5 |
1 |
58 |
314.34 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.46 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8100 |
0.32 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.46 |
Binding ≤ 10μM
|
|
F16P1-1-E |
Fructose-1,6-bisphosphatase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8100 |
0.32 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK12-2-E |
MAP Kinase P38 Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MK14-3-E |
MAP Kinase P38 Alpha (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6300 |
0.33 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
630 |
0.39 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.37 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1489 |
0.37 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Functional ≤ 10μM |
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
143 |
0.44 |
Functional ≤ 10μM
|
|
ERBB4-1-E |
Receptor Protein-tyrosine Kinase ErbB-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.47 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
211 |
0.42 |
Functional ≤ 10μM
|
|
Z100731-1-O |
Bcap37 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
8900 |
0.32 |
Functional ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
4400 |
0.34 |
Functional ≤ 10μM
|
|
Z80037-1-O |
Balb/MK (cluster #1 Of 1), Other |
Other |
200 |
0.43 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4700 |
0.34 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
8900 |
0.32 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
820 |
0.39 |
Functional ≤ 10μM
|
|
Z80852-3-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
8310 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
8.59 |
-10.58 |
1 |
5 |
0 |
56 |
360.211 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
-0.15 |
-10.09 |
1 |
5 |
0 |
56 |
388.265 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
32 |
0.48 |
Binding ≤ 10μM
|
|
VGFR2-2-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.35 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
179 |
0.43 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
179 |
0.43 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1950 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
1.53 |
-8.82 |
1 |
5 |
0 |
56 |
299.305 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACVR1-1-E |
Activin Receptor Type-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.19 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.20 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.18 |
Binding ≤ 10μM
|
|
CDKL2-1-E |
Cyclin-dependent Kinase-like 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.19 |
Binding ≤ 10μM
|
|
CLK1-2-E |
Dual Specificty Protein Kinase CLK1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.21 |
Binding ≤ 10μM
|
|
CLK2-1-E |
Dual Specificity Protein Kinase CLK2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1900 |
0.20 |
Binding ≤ 10μM
|
|
CLK3-1-E |
Dual Specificity Protein Kinase CLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.17 |
Binding ≤ 10μM
|
|
CLK4-1-E |
Dual Specificity Protein Kinase CLK4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.20 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.28 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.20 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
870 |
0.21 |
Binding ≤ 10μM
|
|
EPHB6-1-E |
Ephrin Type-B Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.20 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.27 |
Binding ≤ 10μM
|
|
HIPK4-1-E |
Homeodomain-interacting Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.22 |
Binding ≤ 10μM
|
|
IRAK1-1-E |
Interleukin-1 Receptor-associated Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.19 |
Binding ≤ 10μM
|
|
IRAK3-1-E |
Interleukin-1 Receptor-associated Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
730 |
0.21 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.29 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.21 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.22 |
Binding ≤ 10μM
|
|
NTRK2-1-E |
Neurotrophic Tyrosine Kinase Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.21 |
Binding ≤ 10μM
|
|
NTRK3-1-E |
NT-3 Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.19 |
Binding ≤ 10μM
|
|
PGFRA-2-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.30 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.28 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.18 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.19 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2070 |
0.19 |
Binding ≤ 10μM |
|
Z100405-1-O |
MV4-11 (Myeloid Leukemia Cells) (cluster #1 Of 1), Other |
Other |
60 |
0.25 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
1000 |
0.20 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.63 |
-2.28 |
-49.63 |
2 |
10 |
1 |
93 |
563.723 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.32 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1650 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
8.91 |
-7.7 |
1 |
5 |
0 |
56 |
378.201 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8400 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
7.11 |
-9.1 |
2 |
4 |
0 |
55 |
250.305 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
68 |
0.37 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
309 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.34 |
11.31 |
-9.67 |
1 |
5 |
0 |
56 |
357.413 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
-0.74 |
-11.03 |
1 |
5 |
0 |
55 |
354.211 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.28 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.18 |
Binding ≤ 10μM
|
|
CSK-1-E |
Tyrosine-protein Kinase CSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.22 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.21 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.25 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.31 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.18 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.31 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
76 |
0.26 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
220 |
0.25 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
140 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
9.82 |
-45.05 |
2 |
10 |
1 |
92 |
543.044 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.46 |
9.74 |
-46.9 |
2 |
10 |
1 |
92 |
543.044 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.46 |
7.35 |
-11.91 |
1 |
10 |
0 |
90 |
542.036 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7670 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
1.46 |
-16.23 |
2 |
4 |
0 |
71 |
178.143 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAPK1-1-E |
AMP-activated Protein Kinase, Alpha-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
94 |
0.33 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
69 |
0.33 |
Binding ≤ 10μM
|
|
ACVL1-1-E |
Serine/threonine-protein Kinase Receptor R3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.30 |
Binding ≤ 10μM
|
|
ACVR1-1-E |
Activin Receptor Type-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
150 |
0.32 |
Binding ≤ 10μM
|
|
ADCK3-1-E |
Chaperone Activity Of Bc1 Complex-like, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.25 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.27 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.26 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.27 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.34 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.28 |
Binding ≤ 10μM
|
|
CSK-1-E |
Tyrosine-protein Kinase CSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.25 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.27 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.37 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.30 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.37 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.29 |
Binding ≤ 10μM
|
|
EPHA1-1-E |
Ephrin Type-A Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.31 |
Binding ≤ 10μM
|
|
EPHA2-1-E |
Ephrin Type-A Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.27 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.27 |
Binding ≤ 10μM
|
|
EPHA4-1-E |
Ephrin Type-A Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4300 |
0.25 |
Binding ≤ 10μM
|
|
EPHA5-1-E |
Ephrin Type-A Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.30 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.34 |
Binding ≤ 10μM
|
|
EPHA7-1-E |
Ephrin Type-A Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.26 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
91 |
0.33 |
Binding ≤ 10μM
|
|
EPHB1-1-E |
Ephrin Type-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
540 |
0.29 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.30 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
520 |
0.29 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.26 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
560 |
0.29 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.25 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.25 |
Binding ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.24 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
270 |
0.31 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.28 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.29 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5700 |
0.24 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.33 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4200 |
0.25 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
560 |
0.29 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
240 |
0.31 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
61 |
0.34 |
Binding ≤ 10μM
|
|
LTK-1-E |
Leukocyte Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.29 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.29 |
Binding ≤ 10μM
|
|
M3K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.25 |
Binding ≤ 10μM
|
|
M4K3-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.27 |
Binding ≤ 10μM
|
|
M4K4-2-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.27 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.29 |
Binding ≤ 10μM
|
|
MERTK-1-E |
Proto-oncogene Tyrosine-protein Kinase MER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.27 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.24 |
Binding ≤ 10μM
|
|
MK06-1-E |
Mitogen-activated Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.27 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
360 |
0.30 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.26 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5100 |
0.25 |
Binding ≤ 10μM
|
|
MP2K1-3-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.27 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.28 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.26 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Binding ≤ 10μM
|
|
PGFRA-2-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.31 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
88 |
0.33 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.28 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7100 |
0.24 |
Binding ≤ 10μM
|
|
PHKG2-1-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.23 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.29 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.35 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
97 |
0.33 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.39 |
Binding ≤ 10μM
|
|
SIK1-1-E |
Serine/threonine-protein Kinase SIK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.27 |
Binding ≤ 10μM
|
|
SIK2-1-E |
Serine/threonine-protein Kinase SIK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
430 |
0.30 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
96 |
0.33 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.33 |
Binding ≤ 10μM
|
|
SRMS-1-E |
Tyrosine-protein Kinase Srms (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.27 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
81 |
0.33 |
Binding ≤ 10μM
|
|
STK33-1-E |
Serine/threonine-protein Kinase 33 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.27 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
480 |
0.29 |
Binding ≤ 10μM
|
|
TESK1-1-E |
Dual Specificity Testis-specific Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.25 |
Binding ≤ 10μM
|
|
TIE1-1-E |
Tyrosine-protein Kinase Receptor Tie-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.27 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.25 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.26 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3900 |
0.25 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3700 |
0.25 |
Binding ≤ 10μM
|
|
TYRO3-1-E |
Tyrosine-protein Kinase Receptor TYRO3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
93 |
0.33 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.31 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.31 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.27 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.38 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.35 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.30 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.31 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
170 |
0.32 |
Functional ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.28 |
Functional ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.28 |
Functional ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.28 |
Functional ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.34 |
Functional ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Functional ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.34 |
Functional ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Functional ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.34 |
Functional ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.30 |
Functional ≤ 10μM
|
|
Z100768-1-O |
TPC1 (Papillary Thyroid Carinoma Cells) (cluster #1 Of 1), Other |
Other |
116 |
0.32 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
4200 |
0.25 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
60 |
0.34 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
400 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.98 |
13.02 |
-39.96 |
2 |
6 |
1 |
61 |
476.37 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.30 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.23 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.29 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.34 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.30 |
Functional ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Functional ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Functional ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
200 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
10.77 |
-40.94 |
2 |
7 |
1 |
64 |
446.934 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.97 |
10.65 |
-40.28 |
2 |
7 |
1 |
64 |
446.934 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.97 |
8.3 |
-9.4 |
1 |
7 |
0 |
63 |
445.926 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5470 |
0.32 |
Binding ≤ 10μM
|
|
BMX-1-E |
Tyrosine-protein Kinase BMX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
324 |
0.40 |
Binding ≤ 10μM
|
|
BTK-1-E |
Tyrosine-protein Kinase BTK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5830 |
0.32 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
ERBB4-1-E |
Receptor Protein-tyrosine Kinase ErbB-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.48 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3140 |
0.33 |
Binding ≤ 10μM
|
|
Z80262-1-O |
NCI-H1975 (Bronchoalveolar Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
610 |
0.38 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
15 |
0.48 |
Functional ≤ 10μM
|
|
Z80852-3-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
95 |
0.43 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
4133 |
0.33 |
Functional ≤ 10μM
|
|
Z81331-2-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
8230 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
-1.27 |
-13.95 |
2 |
5 |
0 |
66 |
369.222 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
346 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
0.2 |
-8.57 |
1 |
3 |
0 |
37 |
221.263 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
-0.56 |
-9.48 |
1 |
3 |
0 |
37 |
300.159 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
18 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
-1.89 |
-6 |
1 |
4 |
0 |
50 |
335.592 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
684 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
-0.8 |
-14.3 |
1 |
4 |
0 |
50 |
319.137 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
-1.62 |
-7.46 |
1 |
4 |
0 |
50 |
335.592 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
-3.02 |
-14.4 |
2 |
5 |
0 |
62 |
330.189 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.60 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
6.34 |
-11.7 |
3 |
5 |
0 |
77 |
316.162 |
2 |
↓
|
|
|
|
Analogs
-
32188977
-
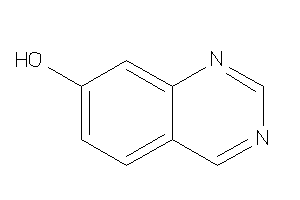
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.93 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
4.01 |
-9.73 |
0 |
3 |
0 |
35 |
160.176 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.32 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7400 |
0.31 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.38 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.46 |
Binding ≤ 10μM
|
|
F16P1-2-E |
Fructose-1,6-bisphosphatase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
INSR-1-E |
Insulin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.34 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8300 |
0.31 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.37 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.34 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.43 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
5700 |
0.32 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.43 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4900 |
0.32 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
6500 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
1.17 |
-9.7 |
1 |
5 |
0 |
56 |
333.75 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
27 |
1.06 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
4.68 |
-7.46 |
0 |
2 |
0 |
26 |
130.15 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
263 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
-2.25 |
-12.78 |
1 |
5 |
0 |
59 |
331.173 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
2.76 |
-11.73 |
1 |
5 |
0 |
56 |
349.312 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
730 |
0.25 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
870 |
0.25 |
Binding ≤ 10μM
|
|
ACVR1-1-E |
Activin Receptor Type-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.24 |
Binding ≤ 10μM
|
|
ADCK3-1-E |
Chaperone Activity Of Bc1 Complex-like, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.24 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.23 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.22 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.22 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.30 |
Binding ≤ 10μM
|
|
BMP2K-1-E |
BMP-2-inducible Protein Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.22 |
Binding ≤ 10μM
|
|
BMX-1-E |
Tyrosine-protein Kinase BMX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
62 |
0.30 |
Binding ≤ 10μM
|
|
BTK-1-E |
Tyrosine-protein Kinase BTK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
750 |
0.25 |
Binding ≤ 10μM
|
|
CSK-1-E |
Tyrosine-protein Kinase CSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.21 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.24 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.26 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.34 |
Binding ≤ 10μM
|
|
EPHA1-1-E |
Ephrin Type-A Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.23 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
EPHA4-1-E |
Ephrin Type-A Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.22 |
Binding ≤ 10μM
|
|
EPHA5-1-E |
Ephrin Type-A Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.27 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
72 |
0.29 |
Binding ≤ 10μM
|
|
EPHA7-1-E |
Ephrin Type-A Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.24 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.25 |
Binding ≤ 10μM
|
|
EPHB1-1-E |
Ephrin Type-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.24 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.23 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.22 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.33 |
Binding ≤ 10μM
|
|
ERBB4-1-E |
Receptor Protein-tyrosine Kinase ErbB-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.31 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.23 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
730 |
0.25 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8100 |
0.21 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.30 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.22 |
Binding ≤ 10μM
|
|
IRAK3-1-E |
Interleukin-1 Receptor-associated Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Binding ≤ 10μM
|
|
ITK-1-E |
Tyrosine-protein Kinase ITK/TSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5650 |
0.22 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.26 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7900 |
0.21 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.21 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.26 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
810 |
0.25 |
Binding ≤ 10μM
|
|
M3K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.21 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.23 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.22 |
Binding ≤ 10μM
|
|
MK06-1-E |
Mitogen-activated Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.24 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.22 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.27 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.24 |
Binding ≤ 10μM
|
|
MP2K1-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.24 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.24 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5200 |
0.22 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7500 |
0.21 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.22 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.25 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.27 |
Binding ≤ 10μM
|
|
SIK2-1-E |
Serine/threonine-protein Kinase SIK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.22 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.26 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
760 |
0.25 |
Binding ≤ 10μM
|
|
SRMS-1-E |
Tyrosine-protein Kinase Srms (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6700 |
0.21 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.22 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
430 |
0.26 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.22 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
29 |
0.31 |
Binding ≤ 10μM
|
|
TEC-1-E |
Tyrosine-protein Kinase TEC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.24 |
Binding ≤ 10μM
|
|
TESK1-1-E |
Dual Specificity Testis-specific Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.25 |
Binding ≤ 10μM
|
|
TIE1-1-E |
Tyrosine-protein Kinase Receptor Tie-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.23 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.22 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.25 |
Binding ≤ 10μM
|
|
TYRO3-1-E |
Tyrosine-protein Kinase Receptor TYRO3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.22 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.23 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.36 |
Functional ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.34 |
Functional ≤ 10μM
|
|
Z80961-1-O |
HN5 (Squamous Cell Carcinoma) (cluster #1 Of 1), Other |
Other |
50 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
8.5 |
-12.22 |
2 |
8 |
0 |
89 |
485.947 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.04 |
2.51 |
-7.92 |
1 |
5 |
0 |
67 |
347.418 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
7.77 |
-8.85 |
1 |
5 |
0 |
56 |
295.342 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.54 |
8.24 |
-32.45 |
2 |
5 |
1 |
58 |
296.35 |
5 |
↓
|
|
|
|
Analogs
-
31949151
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
-0.99 |
-9.12 |
1 |
4 |
0 |
47 |
265.316 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
-0.17 |
-9.47 |
1 |
5 |
0 |
56 |
360.211 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
-4.29 |
-11.02 |
5 |
5 |
0 |
89 |
330.189 |
2 |
↓
|
|
|
|
Analogs
-
26967
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
55 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
0.23 |
-7.96 |
1 |
4 |
0 |
47 |
251.289 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
-2.03 |
-8.98 |
3 |
4 |
0 |
63 |
270.723 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
10.05 |
-8.32 |
1 |
6 |
0 |
84 |
345.156 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.57 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
0.05 |
-9.53 |
1 |
3 |
0 |
37 |
255.708 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
-1.84 |
-9.92 |
3 |
4 |
0 |
63 |
236.278 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
508 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
6.42 |
-8.04 |
3 |
4 |
0 |
64 |
250.305 |
3 |
↓
|
|
|
|
Analogs
-
31949151
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
-1.01 |
-7.3 |
1 |
4 |
0 |
47 |
265.316 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.44 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6390 |
0.38 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
7200 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
-1.52 |
-8.88 |
3 |
4 |
0 |
63 |
250.305 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.49 |
Binding ≤ 10μM |
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3290 |
0.40 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
4930 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
7.17 |
-9.8 |
3 |
4 |
0 |
64 |
270.723 |
2 |
↓
|
|
|
|
Analogs
-
36892190
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
35 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
-1.29 |
-7.31 |
1 |
3 |
0 |
37 |
306.188 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5500 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.95 |
7.89 |
-7.32 |
1 |
2 |
0 |
25 |
299.171 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
-2.64 |
-8.9 |
3 |
4 |
0 |
63 |
315.174 |
2 |
↓
|
|
|
|
Analogs
-
41480387
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
910 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
9.44 |
-8.45 |
1 |
3 |
0 |
38 |
235.29 |
2 |
↓
|
|
|
|
Analogs
-
44124360
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.24 |
Binding ≤ 10μM
|
|
CDK8-1-E |
Cell Division Protein Kinase 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.23 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
720 |
0.25 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.24 |
Binding ≤ 10μM
|
|
TBA1A-2-E |
Tubulin Alpha-3 Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
260 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
-1.01 |
-20.16 |
2 |
8 |
0 |
98 |
469.545 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
324 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
-0.94 |
-14.42 |
1 |
5 |
0 |
53 |
344.216 |
3 |
↓
|
|