|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.45 |
10.61 |
-9.06 |
2 |
4 |
0 |
67 |
378.553 |
18 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
33822153
-
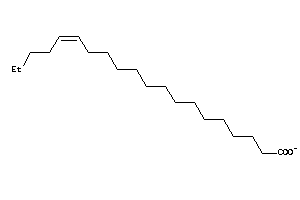
-
4529321
-
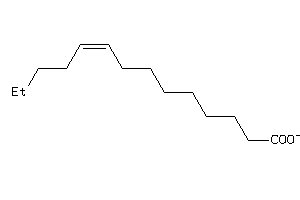
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
FABP4-2-E |
Fatty Acid Binding Protein Adipocyte (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
185 |
0.47 |
Binding ≤ 10μM
|
|
FABP5-2-E |
Fatty Acid Binding Protein Epidermal (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
248 |
0.46 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.58 |
13.34 |
-45.08 |
0 |
2 |
-1 |
40 |
281.46 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EST1-5-E |
Carboxylesterase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
6 |
0.72 |
Binding ≤ 10μM
|
|
EST2-2-E |
Carboxylesterase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.70 |
Binding ≤ 10μM
|
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.57 |
Binding ≤ 10μM
|
|
Q9GPG0-1-E |
Juvenile Hormone Esterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
9.25 |
-5.09 |
0 |
1 |
0 |
17 |
256.333 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.00 |
13.37 |
-8.25 |
0 |
3 |
0 |
39 |
336.516 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.00 |
13.37 |
-8.28 |
0 |
3 |
0 |
39 |
336.516 |
17 |
↓
|
|
|
|
Analogs
-
43569509
-
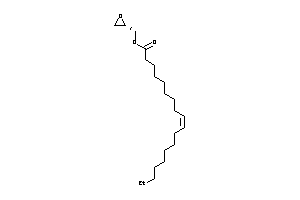
-
43569511
-
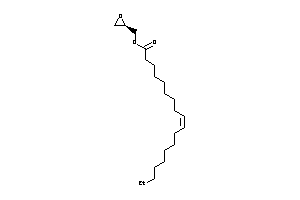
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.71 |
12.14 |
-7.4 |
0 |
3 |
0 |
39 |
310.478 |
16 |
↓
|
|
|
|
Analogs
-
43569509
-

-
43569511
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
8200 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.71 |
12.14 |
-7.43 |
0 |
3 |
0 |
39 |
310.478 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
MGLL-5-E |
Monoglyceride Lipase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
4500 |
0.29 |
Binding ≤ 10μM
|
|
MGLL-5-E |
Monoglyceride Lipase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
6300 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.56 |
14.33 |
-8.78 |
0 |
3 |
0 |
39 |
360.538 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.98 |
13.07 |
-8.52 |
0 |
3 |
0 |
39 |
334.5 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
1600 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.98 |
13.07 |
-8.54 |
0 |
3 |
0 |
39 |
334.5 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-2-E |
Cannabinoid CB1 Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
480 |
0.33 |
Binding ≤ 10μM |
|
CNR2-2-E |
Cannabinoid CB2 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.40 |
Functional ≤ 10μM
|
|
CNR2-2-E |
Cannabinoid CB2 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.45 |
11.89 |
-8.62 |
2 |
4 |
0 |
67 |
378.553 |
18 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.45 |
10.61 |
-9.04 |
2 |
4 |
0 |
67 |
378.553 |
18 |
↓
|
|