|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.64 |
12.98 |
-7.68 |
0 |
5 |
0 |
64 |
379.541 |
9 |
↓
|
|
|
|
Analogs
-
4073970
-
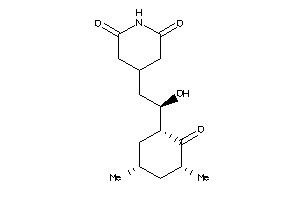
-
4536598
-
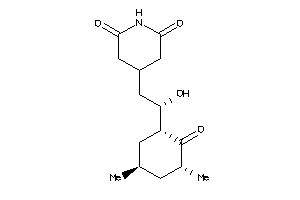
-
8579261
-
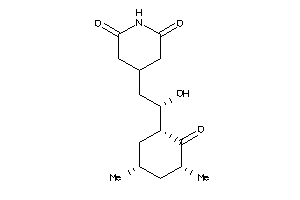
-
11592912
-
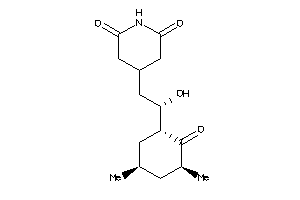
-
11592913
-
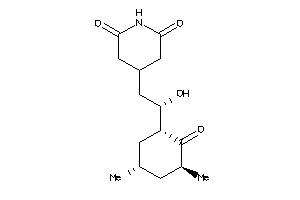
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.38 |
Binding ≤ 10μM
|
|
Z50347-2-O |
Saccharomyces Cerevisiae (cluster #2 Of 3), Other |
Other |
88 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
400 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z80026-2-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
3600 |
0.38 |
Functional ≤ 10μM |
|
Z80224-6-O |
MCF7 (Breast Carcinoma Cells) (cluster #6 Of 14), Other |
Other |
1500 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3500 |
0.38 |
Functional ≤ 10μM
|
|
Z80485-2-O |
SK-MEL-28 (Melanoma Cells) (cluster #2 Of 6), Other |
Other |
1000 |
0.42 |
Functional ≤ 10μM
|
|
Z80490-1-O |
SK-MES-1 (cluster #1 Of 4), Other |
Other |
2700 |
0.39 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 7), Other |
Other |
530 |
0.44 |
Functional ≤ 10μM
|
|
Z80653-1-O |
9L (Glioma Cells) (cluster #1 Of 2), Other |
Other |
200 |
0.47 |
Functional ≤ 10μM
|
|
Z80874-4-O |
CEM (T-cell Leukemia) (cluster #4 Of 7), Other |
Other |
80 |
0.50 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
2500 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-7-O |
Jurkat (Acute Leukemic T-cells) (cluster #7 Of 10), Other |
Other |
930 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-3-O |
LNCaP (Prostate Carcinoma) (cluster #3 Of 5), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM
|
|
Z81247-6-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #6 Of 9), Other |
Other |
533 |
0.44 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM |
|
Z80005-2-O |
3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
912 |
0.42 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
3.22 |
-17 |
2 |
5 |
0 |
83 |
281.352 |
3 |
↓
|
|
|
|
Analogs
-
13446278
-
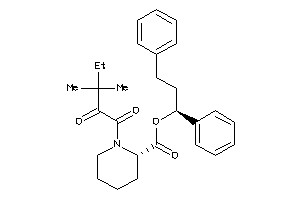
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.33 |
Binding ≤ 10μM
|
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.53 |
3.47 |
-8.6 |
0 |
5 |
0 |
64 |
449.591 |
10 |
↓
|
|
|
|
Analogs
-
38444630
-
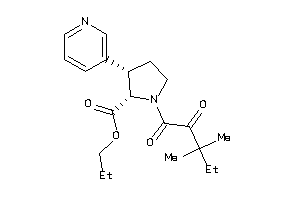
-
38444631
-
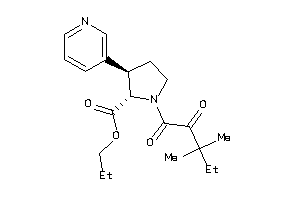
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
2.36 |
-12.12 |
0 |
6 |
0 |
76 |
360.454 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.21 |
2.48 |
-40.9 |
1 |
6 |
1 |
77 |
361.462 |
9 |
↓
|
|
|
|
Analogs
-
38444630
-

-
38444631
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
2.35 |
-12.14 |
0 |
6 |
0 |
76 |
360.454 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.21 |
2.47 |
-40.96 |
1 |
6 |
1 |
77 |
361.462 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
9.05 |
-11.92 |
0 |
6 |
0 |
73 |
305.33 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
9.05 |
-11.98 |
0 |
6 |
0 |
73 |
305.33 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.37 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
7.29 |
-11.51 |
0 |
5 |
0 |
64 |
311.403 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.37 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
7.29 |
-11.5 |
0 |
5 |
0 |
64 |
311.403 |
6 |
↓
|
|
|
|
Analogs
-
39654816
-
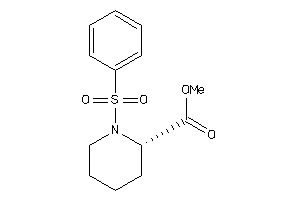
-
6584065
-
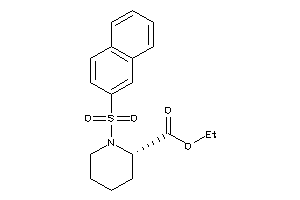
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
6.67 |
-10.77 |
0 |
5 |
0 |
64 |
297.376 |
5 |
↓
|
|
|
|
Analogs
-
39654816
-

-
6584065
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
6.63 |
-10.67 |
0 |
5 |
0 |
64 |
297.376 |
5 |
↓
|
|
|
|
Analogs
-
10027147
-
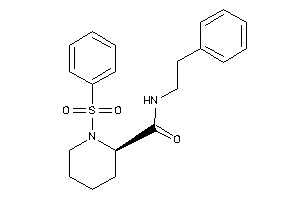
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
8.27 |
-13 |
1 |
5 |
0 |
66 |
372.49 |
6 |
↓
|
|
|
|
Analogs
-
25426373
-
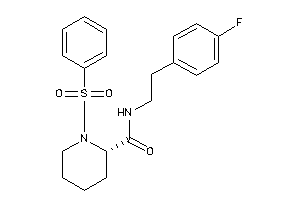
-
25426377
-
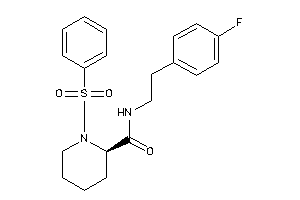
-
10027145
-
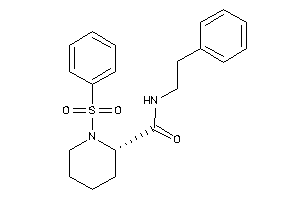
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
8.22 |
-13.21 |
1 |
5 |
0 |
66 |
372.49 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
9.05 |
-12.31 |
1 |
5 |
0 |
66 |
386.517 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
9 |
-12.39 |
1 |
5 |
0 |
66 |
386.517 |
7 |
↓
|
|
|
|
Analogs
-
9095354
-
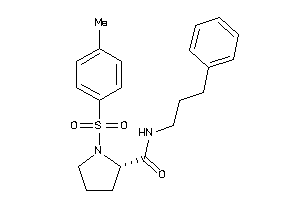
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7600 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
9.16 |
-19.85 |
1 |
5 |
0 |
66 |
386.517 |
7 |
↓
|
|
|
|
Analogs
-
9095353
-
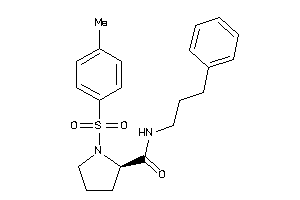
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7600 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
8.5 |
-19.46 |
1 |
5 |
0 |
66 |
386.517 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
10368169
-
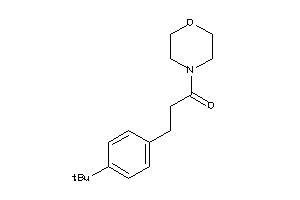
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
0.44 |
-7.68 |
0 |
3 |
0 |
29 |
261.365 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FKB1A-1-E |
FK506-binding Protein 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM
|
|
Q8QGU2-1-E |
FK506 Binding Protein 12 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.41 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
8.24 |
-8.13 |
0 |
5 |
0 |
64 |
283.368 |
6 |
↓
|
|