|
|
Analogs
-
4245675
-
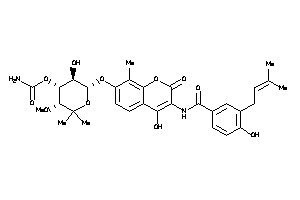
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
4.17 |
-59.66 |
5 |
13 |
-1 |
203 |
611.624 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.93 |
3.05 |
-27.62 |
6 |
13 |
0 |
200 |
612.632 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.93 |
3.81 |
-30.74 |
6 |
13 |
0 |
200 |
612.632 |
9 |
↓
|
|
|
|
|
|
|
Analogs
-
22063165
-
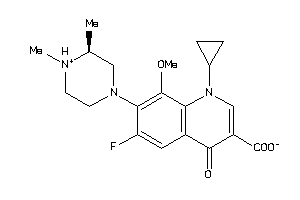
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.04 |
8.97 |
-101.63 |
2 |
7 |
0 |
91 |
375.4 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.04 |
7.73 |
-63.01 |
1 |
7 |
-1 |
87 |
374.392 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.78 |
7.72 |
-78.04 |
3 |
7 |
1 |
94 |
376.408 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-0.04 |
7.83 |
-63.43 |
1 |
7 |
-1 |
87 |
374.392 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.04 |
7.05 |
-52.32 |
3 |
7 |
1 |
88 |
376.408 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.78 |
7.73 |
-77.5 |
3 |
7 |
1 |
94 |
376.408 |
3 |
↓
|
|
|
|
Analogs
-
22059930
-

-
11616796
-
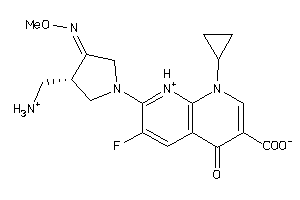
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.32 |
Binding ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.32 |
Binding ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
300 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.76 |
8.21 |
-103.97 |
3 |
9 |
0 |
128 |
389.387 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.76 |
7.85 |
-62.36 |
2 |
9 |
-1 |
126 |
388.379 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.30 |
6.82 |
-80.57 |
4 |
9 |
1 |
130 |
390.395 |
4 |
↓
|
|
|
|
Analogs
-
22059926
-
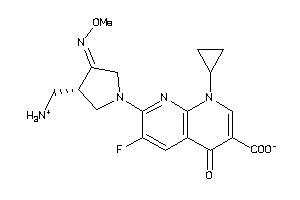
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.32 |
Binding ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.32 |
Binding ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
300 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.76 |
8.24 |
-101.68 |
3 |
9 |
0 |
128 |
389.387 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.76 |
7.69 |
-64.02 |
2 |
9 |
-1 |
126 |
388.379 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.30 |
6.86 |
-79.66 |
4 |
9 |
1 |
130 |
390.395 |
4 |
↓
|
|
|
|
Analogs
-
597112
-
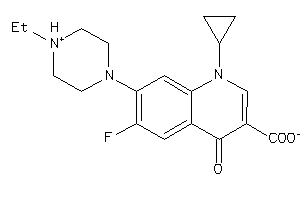
-
6385314
-
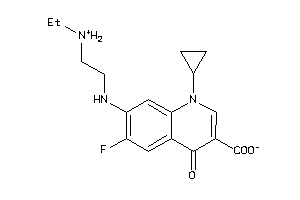
-
39294843
-
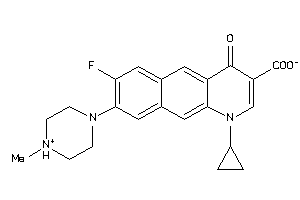
Draw
Identity
99%
90%
80%
70%
Vendors
And 85 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.37 |
Binding ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
500 |
0.37 |
Binding ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
2300 |
0.33 |
Binding ≤ 10μM
|
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
200 |
0.39 |
Functional ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
200 |
0.39 |
Functional ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
2500 |
0.33 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3020 |
0.32 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.44 |
Binding ≤ 10μM |
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.44 |
Binding ≤ 10μM |
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
410 |
0.37 |
Binding ≤ 10μM
|
|
Z50117-1-O |
Pseudomonas Aeruginosa (cluster #1 Of 2), Other |
Other |
220 |
0.39 |
Functional ≤ 10μM
|
|
Z50185-3-O |
Staphylococcus Aureus (cluster #3 Of 4), Other |
Other |
400 |
0.37 |
Functional ≤ 10μM
|
|
Z50212-1-O |
Escherichia Coli (cluster #1 Of 7), Other |
Other |
10 |
0.47 |
Functional ≤ 10μM
|
|
Z50378-1-O |
Mycobacterium Intracellulare (cluster #1 Of 1), Other |
Other |
910 |
0.35 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
1980 |
0.33 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9440 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.70 |
9.07 |
-109.14 |
2 |
6 |
0 |
82 |
331.347 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.70 |
7.77 |
-65.4 |
1 |
6 |
-1 |
77 |
330.339 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.45 |
7.06 |
-88.49 |
3 |
6 |
1 |
85 |
332.355 |
2 |
↓
|
|
|
|
Analogs
-
1894
-
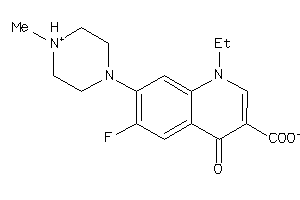
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GYRA-1-B |
DNA Gyrase Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
600 |
0.38 |
Binding ≤ 10μM
|
|
GYRB-1-B |
DNA Gyrase Subunit B (cluster #1 Of 1), Bacterial |
Bacteria |
600 |
0.38 |
Binding ≤ 10μM
|
|
PARC-1-B |
Topoisomerase IV Subunit A (cluster #1 Of 1), Bacterial |
Bacteria |
3500 |
0.33 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
20 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.69 |
8.56 |
-108.13 |
2 |
6 |
0 |
82 |
319.336 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
7.25 |
-65.36 |
1 |
6 |
-1 |
77 |
318.328 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.43 |
6.54 |
-88.24 |
3 |
6 |
1 |
85 |
320.344 |
2 |
↓
|
|