|
|
Analogs
-
1532560
-
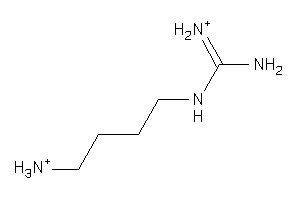
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDE4-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
|
NMDZ1-5-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
9130 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.42 |
0.75 |
-75.07 |
10 |
6 |
2 |
127 |
174.252 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-3-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE2-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7090 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.94 |
2.96 |
-76.97 |
10 |
6 |
2 |
130 |
222.296 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE4-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDZ1-5-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
Z50418-3-O |
Trypanosoma Brucei (cluster #3 Of 6), Other |
Other |
1000 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
-2.06 |
-43 |
3 |
1 |
1 |
27 |
194.342 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
790 |
0.57 |
Binding ≤ 10μM
|
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
790 |
0.57 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
790 |
0.57 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
790 |
0.57 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
790 |
0.57 |
Binding ≤ 10μM
|
|
NMDE4-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
790 |
0.57 |
Binding ≤ 10μM
|
|
NMDZ1-5-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
790 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
5.89 |
-45.85 |
3 |
1 |
1 |
28 |
208.369 |
2 |
↓
|
|
|
|
Analogs
-
3886053
-
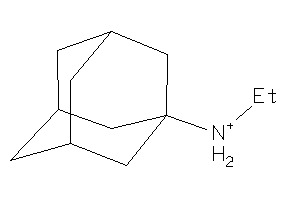
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1720 |
0.54 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1720 |
0.54 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1720 |
0.54 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1720 |
0.54 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1720 |
0.54 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1720 |
0.54 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1720 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.52 |
7.17 |
-35.43 |
2 |
1 |
1 |
17 |
208.369 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
13777648
-
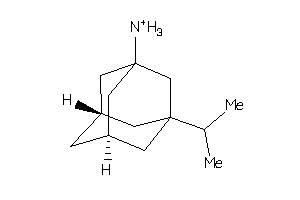
-
31771167
-
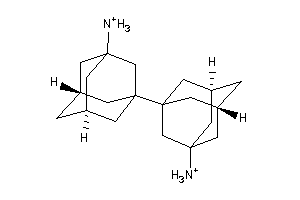
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7210 |
0.51 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7210 |
0.51 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7210 |
0.51 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7210 |
0.51 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7210 |
0.51 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7210 |
0.51 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7210 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
-1.43 |
-42.43 |
3 |
1 |
1 |
27 |
194.342 |
1 |
↓
|
|
|
|
Analogs
-
31771168
-
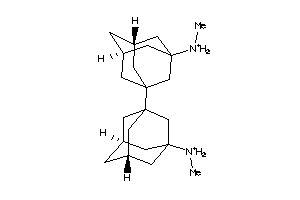
-
2379538
-
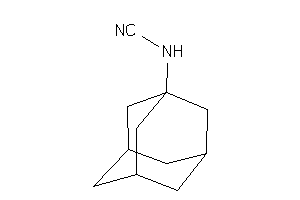
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1610 |
0.58 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1610 |
0.58 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1610 |
0.58 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1610 |
0.58 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1610 |
0.58 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1610 |
0.58 |
Binding ≤ 10μM
|
|
NMDZ1-5-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1610 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
6.36 |
-36.94 |
2 |
1 |
1 |
17 |
194.342 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
|
NMDE4-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
|
NMDZ1-5-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
-2.22 |
-44.3 |
3 |
1 |
1 |
27 |
180.315 |
2 |
↓
|
|
|
|
Analogs
-
968256
-
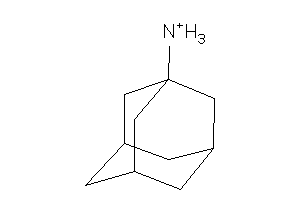
Draw
Identity
99%
90%
80%
70%
Vendors
And 72 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
9520 |
0.54 |
Binding ≤ 10μM
|
|
Z104297-3-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #3 Of 3), Other |
Other |
5500 |
0.57 |
Binding ≤ 10μM
|
|
Z104298-2-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #2 Of 2), Other |
Other |
5600 |
0.57 |
Binding ≤ 10μM
|
|
Z104297-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #1 Of 1), Other |
Other |
1020 |
0.65 |
Functional ≤ 10μM
|
|
Z104298-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #1 Of 1), Other |
Other |
911 |
0.65 |
Functional ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
1585 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
4.76 |
-42.23 |
3 |
1 |
1 |
28 |
180.315 |
0 |
↓
|
|