|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-3-E |
Cyclin A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.35 |
Binding ≤ 10μM
|
|
CDK4-1-E |
Cyclin-dependent Kinase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
DYR1A-3-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.30 |
Binding ≤ 10μM
|
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
40 |
0.37 |
Binding ≤ 10μM
|
|
PDK1-1-E |
Pyruvate Dehydrogenase Kinase Isoform 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.30 |
Binding ≤ 10μM
|
|
PDPK1-1-E |
3-phosphoinositide Dependent Protein Kinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
ROCK2-1-E |
Rho-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
5.84 |
-17.28 |
4 |
9 |
0 |
136 |
402.48 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.30 |
6.38 |
-43.75 |
5 |
9 |
1 |
137 |
403.488 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.04 |
5.48 |
-18.48 |
4 |
9 |
0 |
139 |
402.48 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5400 |
0.20 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.20 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.24 |
Binding ≤ 10μM
|
|
PDK1-1-E |
Pyruvate Dehydrogenase Kinase Isoform 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4300 |
0.20 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.21 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
11.99 |
-24.02 |
1 |
9 |
0 |
96 |
494.555 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
AURKA-2-E |
Serine/threonine-protein Kinase Aurora-A (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
JAK3-2-E |
Tyrosine-protein Kinase JAK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
KS6A3-1-E |
Ribosomal Protein S6 Kinase Alpha 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
MERTK-1-E |
Proto-oncogene Tyrosine-protein Kinase MER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
PDK1-1-E |
Pyruvate Dehydrogenase Kinase Isoform 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
ST17A-1-E |
Serine/threonine-protein Kinase 17A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
TYK2-1-E |
Tyrosine-protein Kinase TYK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
2.15 |
-10.32 |
4 |
9 |
0 |
111 |
381.44 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.23 |
4.38 |
-50.13 |
5 |
9 |
1 |
112 |
382.448 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.23 |
4.72 |
-95.17 |
6 |
9 |
2 |
113 |
383.456 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.23 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.24 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.24 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2300 |
0.24 |
Binding ≤ 10μM
|
|
FAK1-1-E |
Focal Adhesion Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8300 |
0.22 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.22 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
870 |
0.26 |
Binding ≤ 10μM
|
|
JAK1-1-E |
Tyrosine-protein Kinase JAK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7300 |
0.22 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Binding ≤ 10μM
|
|
KPCA-5-E |
Protein Kinase C Alpha (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.37 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.35 |
Binding ≤ 10μM
|
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.35 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5400 |
0.22 |
Binding ≤ 10μM
|
|
PDK1-1-E |
Pyruvate Dehydrogenase Kinase Isoform 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6700 |
0.22 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9100 |
0.21 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3900 |
0.23 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
-4.56 |
-45.41 |
3 |
8 |
1 |
99 |
439.499 |
3 |
↓
|
|
|
|
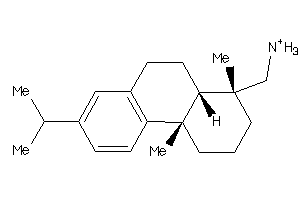
Analogs
-
3947435
-
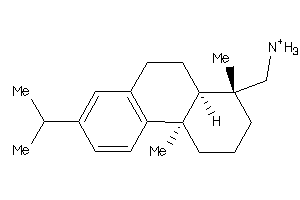
-
3947436
-
-
3947437
-
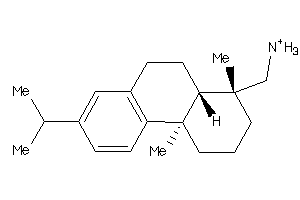
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
8000 |
0.34 |
Binding ≤ 10μM |
|
PDK1-1-E |
Pyruvate Dehydrogenase Kinase Isoform 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9500 |
0.33 |
Binding ≤ 10μM
|
|
PDK2-1-E |
Pyruvate Dehydrogenase Kinase Isoform 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.33 |
Binding ≤ 10μM
|
|
PDK3-1-E |
Pyruvate Dehydrogenase Kinase Isoform 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.33 |
Binding ≤ 10μM
|
|
PDK4-1-E |
Pyruvate Dehydrogenase Kinase Isoform 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.23 |
9.42 |
-46.88 |
3 |
1 |
1 |
28 |
286.483 |
2 |
↓
|
|