|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.74 |
-6.79 |
-52.29 |
4 |
9 |
-1 |
150 |
252.21 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.74 |
-6.65 |
-56.92 |
5 |
9 |
0 |
152 |
253.218 |
4 |
↓
|
|
|
|
Analogs
-
25761394
-
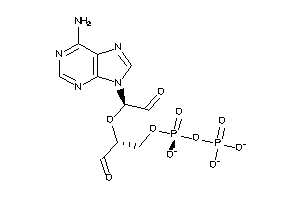
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.63 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
-4.14 |
-16.66 |
3 |
9 |
0 |
133 |
265.229 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.63 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
-4.16 |
-16.97 |
3 |
9 |
0 |
133 |
265.229 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.39 |
Binding ≤ 10μM
|
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.63 |
Binding ≤ 10μM |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
-4.14 |
-17.12 |
3 |
9 |
0 |
133 |
265.229 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.38 |
Binding ≤ 10μM
|
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.63 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
-4.14 |
-17.04 |
3 |
9 |
0 |
133 |
265.229 |
6 |
↓
|
|
|
|
Analogs
-
21290121
-
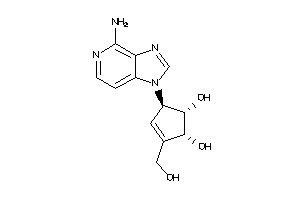
-
34422831
-
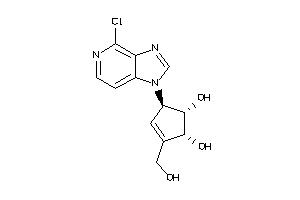
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.55 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-4 |
-13.15 |
5 |
7 |
0 |
117 |
262.269 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.61 |
-3.72 |
-33.27 |
6 |
7 |
1 |
119 |
263.277 |
2 |
↓
|
|
|
|
Analogs
-
34422831
-

-
21290121
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.55 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-3.92 |
-17.2 |
5 |
7 |
0 |
117 |
262.269 |
2 |
↓
|
|
|
|
Analogs
-
34422831
-

-
21290121
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.55 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-2.96 |
-17.44 |
5 |
7 |
0 |
117 |
262.269 |
2 |
↓
|
|
|
|
Analogs
-
34422831
-

-
21290121
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.55 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-2.93 |
-17.91 |
5 |
7 |
0 |
117 |
262.269 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.12 |
-2.04 |
-15.96 |
5 |
10 |
0 |
148 |
294.271 |
2 |
↓
|
|
|
|
Analogs
-
370776
-
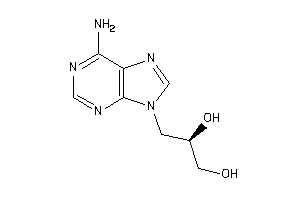
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8500 |
0.47 |
Binding ≤ 10μM
|
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.61 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-1.27 |
-15 |
4 |
7 |
0 |
110 |
209.209 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-5.39 |
-36.69 |
5 |
7 |
1 |
111 |
210.217 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-0.9 |
-38.73 |
5 |
7 |
1 |
111 |
210.217 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6260 |
0.38 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2150 |
0.42 |
Binding ≤ 10μM
|
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4850 |
0.39 |
Binding ≤ 10μM
|
|
Z50467-1-O |
Trichomonas Vaginalis (cluster #1 Of 3), Other |
Other |
10000 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.13 |
-9.63 |
-16.95 |
5 |
8 |
0 |
130 |
265.273 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.39 |
-9.47 |
-36.8 |
6 |
8 |
1 |
132 |
266.281 |
2 |
↓
|
|
|
|
Analogs
-
4513750
-
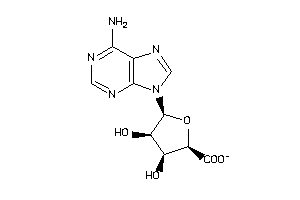
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.98 |
-1.52 |
-44.95 |
4 |
10 |
-1 |
159 |
280.22 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.98 |
-1.26 |
-52.28 |
5 |
10 |
0 |
161 |
281.228 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.98 |
-5.63 |
-54.42 |
5 |
10 |
0 |
161 |
281.228 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.44 |
Binding ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
4600 |
0.37 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
1400 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.29 |
-8.66 |
-19.17 |
5 |
8 |
0 |
130 |
281.247 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.49 |
-8.5 |
-35.66 |
6 |
8 |
1 |
132 |
282.255 |
2 |
↓
|
|
|
|
|