|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-2-E |
Alpha-1a Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
510 |
0.63 |
Binding ≤ 10μM
|
|
ADA1B-4-E |
Alpha-1b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
510 |
0.63 |
Binding ≤ 10μM
|
|
ADA1D-2-E |
Alpha-1d Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
510 |
0.63 |
Binding ≤ 10μM
|
|
ADA2A-2-E |
Alpha-2a Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Binding ≤ 10μM
|
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.81 |
Binding ≤ 10μM
|
|
ADA2C-2-E |
Alpha-2c Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.82 |
Binding ≤ 10μM
|
|
NISCH-1-E |
Nischarin (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
ADA1A-2-E |
Alpha-1a Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.65 |
Functional ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.65 |
Functional ≤ 10μM
|
|
ADA1D-4-E |
Alpha-1d Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
290 |
0.65 |
Functional ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Functional ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Functional ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.84 |
Functional ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
528 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
5.66 |
-7.16 |
2 |
3 |
0 |
36 |
230.098 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-2-E |
Alpha-2a Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.79 |
Binding ≤ 10μM
|
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.79 |
Binding ≤ 10μM
|
|
ADA2C-2-E |
Alpha-2c Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.79 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
5100 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
0.24 |
-32.52 |
5 |
5 |
1 |
78 |
194.214 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
-
- ZP-BB-AS004416
- ZP-BB-AS004424
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
7.21 |
-31.75 |
2 |
3 |
1 |
29 |
212.276 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.40 |
-0.5 |
-37.56 |
2 |
3 |
1 |
31 |
210.26 |
2 |
↓
|
|
|
|
Analogs
-
5626
-
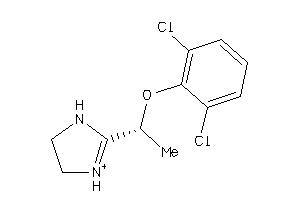
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-2-E |
Alpha-2a Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
|
ADA2C-2-E |
Alpha-2c Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
660 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
-2.62 |
-26.83 |
2 |
3 |
1 |
35 |
260.144 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.49 |
-8.18 |
-17.25 |
6 |
11 |
0 |
175 |
424.443 |
8 |
↓
|
|
|
|
Analogs
-
1266
-
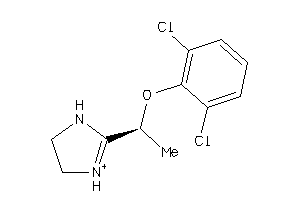
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-2-E |
Alpha-2a Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
|
ADA2C-2-E |
Alpha-2c Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.71 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
660 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
-2.62 |
-26.85 |
2 |
3 |
1 |
35 |
260.144 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
6.41 |
-48.01 |
4 |
8 |
0 |
131 |
442.475 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-4-E |
Alpha-2a Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.87 |
Binding ≤ 10μM
|
|
ADA2B-3-E |
Alpha-2b Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.87 |
Binding ≤ 10μM
|
|
ADA2C-4-E |
Alpha-2c Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.87 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
4850 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
5.13 |
-25.11 |
3 |
3 |
1 |
38 |
216.717 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.50 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.42 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
758 |
0.37 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
55 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
64 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.56 |
-6.67 |
1 |
4 |
0 |
35 |
326.831 |
1 |
↓
|
|
|
|
Analogs
-
12503139
-
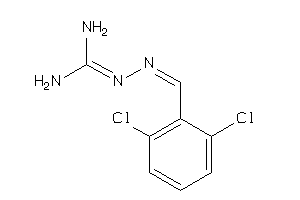
-
34040487
-
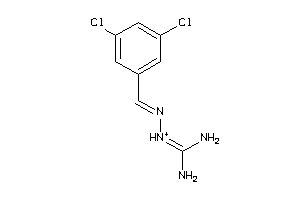
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
5.56 |
-28.39 |
5 |
4 |
1 |
78 |
232.094 |
2 |
↓
|
|
|
|
Analogs
-
13470054
-
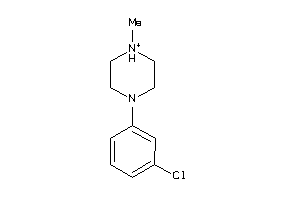
Draw
Identity
99%
90%
80%
70%
Vendors
And 83 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.77 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.77 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
32 |
0.81 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.87 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
62 |
0.78 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
62 |
0.78 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
62 |
0.78 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
62 |
0.78 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
62 |
0.78 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8700 |
0.54 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.61 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
304 |
0.70 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.61 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
6150 |
0.56 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
75 |
0.77 |
Functional ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
287 |
0.70 |
Functional ≤ 10μM
|
|
5HT2C-2-E |
5-hydroxytryptamine Receptor 2C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
26 |
0.82 |
Functional ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
236 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
4.99 |
-43.48 |
2 |
2 |
1 |
20 |
197.689 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.99 |
3.68 |
-3.14 |
1 |
2 |
0 |
15 |
196.681 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 57 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
372 |
0.43 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.54 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
52 |
0.49 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
27 |
0.50 |
Binding ≤ 10μM |
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
27 |
0.50 |
Binding ≤ 10μM |
|
ADCY2-2-E |
Adenylate Cyclase Type II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
ADCY3-2-E |
Adenylate Cyclase Type III (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
ADCY4-2-E |
Adenylate Cyclase Type IV (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
ADCY5-2-E |
Adenylate Cyclase Type V (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
ADCY6-2-E |
Adenylate Cyclase Type VI (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
ADCY8-1-E |
Adenylate Cyclase Type VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
CALM-2-E |
Calmodulin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7260 |
0.34 |
Binding ≤ 10μM
|
|
D4A3N4-1-E |
Brain Adenylate Cyclase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
118 |
0.46 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
34 |
0.50 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.53 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.53 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM
|
|
KCNH2-2-E |
HERG (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5830 |
0.35 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
NMDE1-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
NMDZ1-6-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
600 |
0.41 |
Binding ≤ 10μM
|
|
PRIO-1-E |
Prion Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
|
Q8CFM9-1-E |
Adenylate Cyclase Type VII (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.47 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.36 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.36 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.36 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.36 |
Binding ≤ 10μM
|
|
TRPV1-2-E |
Vanilloid Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Binding ≤ 10μM
|
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5800 |
0.35 |
Binding ≤ 10μM
|
|
ADCY2-1-E |
Adenylate Cyclase Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
ADCY3-1-E |
Adenylate Cyclase Type III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
ADCY4-1-E |
Adenylate Cyclase Type IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
ADCY5-1-E |
Adenylate Cyclase Type V (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
ADCY6-1-E |
Adenylate Cyclase Type VI (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
ADCY8-1-E |
Adenylate Cyclase Type VIII (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
D4A3N4-1-E |
Brain Adenylate Cyclase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1470 |
0.39 |
Functional ≤ 10μM
|
|
Q8CFM9-1-E |
Adenylate Cyclase Type VII (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.39 |
Functional ≤ 10μM
|
|
TRPV1-1-E |
Vanilloid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
520 |
0.42 |
Functional ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.34 |
ADME/T ≤ 10μM
|
|
DRD2-14-E |
Dopamine D2 Receptor (cluster #14 Of 24), Eukaryotic |
Eukaryotes |
10 |
0.53 |
Binding ≤ 10μM
|
|
PDR5-1-F |
Pleiotropic ABC Efflux Transporter Of Multiple Drugs (cluster #1 Of 1), Fungal |
Fungi |
4500 |
0.36 |
Binding ≤ 10μM
|
|
Z104302-1-O |
Glutamate NMDA Receptor (cluster #1 Of 7), Other |
Other |
1300 |
0.39 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
61 |
0.48 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
6 |
0.55 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
51 |
0.49 |
Binding ≤ 10μM
|
|
Z101682-1-O |
BK Polyomavirus (cluster #1 Of 2), Other |
Other |
7600 |
0.34 |
Functional ≤ 10μM
|
|
Z102380-3-O |
Ileum (cluster #3 Of 3), Other |
Other |
180 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-2-O |
Plasmodium Falciparum (cluster #2 Of 22), Other |
Other |
3981 |
0.36 |
Functional ≤ 10μM
|
|
Z50597-9-O |
Rattus Norvegicus (cluster #9 Of 12), Other |
Other |
180 |
0.45 |
Functional ≤ 10μM
|
|
Z50636-3-O |
Sindbis Virus (cluster #3 Of 3), Other |
Other |
8200 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
11.52 |
-43.38 |
1 |
2 |
1 |
9 |
319.881 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM |
|
ADA1B-4-E |
Alpha-1b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM |
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.62 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
52 |
0.60 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.62 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Functional ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
55 |
0.60 |
Functional ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Functional ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
740 |
0.50 |
Binding ≤ 10μM
|
|
Z50425-7-O |
Plasmodium Falciparum (cluster #7 Of 22), Other |
Other |
7943 |
0.42 |
Functional ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 8), Other |
Other |
1 |
0.74 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
3.74 |
-13.23 |
2 |
5 |
0 |
62 |
292.14 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.86 |
4.31 |
-33.95 |
3 |
5 |
1 |
64 |
293.148 |
2 |
↓
|
|
|
|
Analogs
-
31945367
-
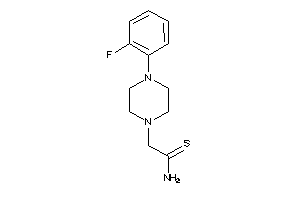
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.78 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.78 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.78 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
58 |
0.78 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
373 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
3.34 |
-39.29 |
2 |
2 |
1 |
20 |
181.234 |
1 |
↓
|
|
|
|
Analogs
-
1556646
-
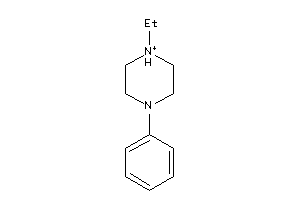
-
8462937
-
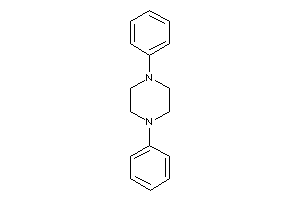
-
34320168
-
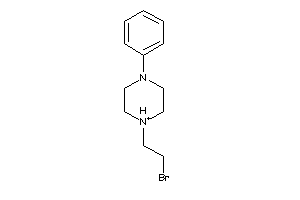
-
39280325
-
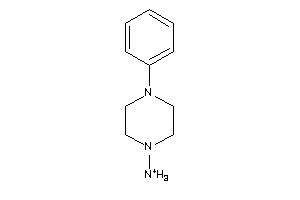
Draw
Identity
99%
90%
80%
70%
Vendors
And 81 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.74 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
63 |
0.84 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
149 |
0.80 |
Binding ≤ 10μM
|
|
5HT1E-2-E |
Serotonin 1e (5-HT1e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
135 |
0.80 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
149 |
0.80 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
668 |
0.72 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
668 |
0.72 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
668 |
0.72 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.64 |
Binding ≤ 10μM
|
|
DRD2-11-E |
Dopamine D2 Receptor (cluster #11 Of 24), Eukaryotic |
Eukaryotes |
6800 |
0.60 |
Binding ≤ 10μM
|
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
794 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
3.36 |
-41.04 |
2 |
2 |
1 |
20 |
163.244 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
6.26 |
-6.75 |
2 |
3 |
0 |
36 |
263.65 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304-2-O |
Adrenergic Receptor Alpha-1 (cluster #2 Of 3), Other |
Other |
263 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
5.99 |
-34.19 |
3 |
3 |
1 |
42 |
243.117 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.68 |
6 |
-34.35 |
3 |
3 |
1 |
42 |
243.117 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.68 |
5.55 |
-8.4 |
2 |
3 |
0 |
41 |
242.109 |
3 |
↓
|
|