|
|
Analogs
-
6846023
-
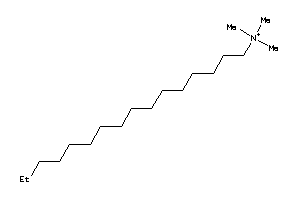
-
1532339
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
892 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
6.92 |
-67.7 |
0 |
2 |
2 |
0 |
244.467 |
10 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYN1-1-E |
Dynamin-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8150 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
900 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
5.16 |
-28.94 |
0 |
1 |
1 |
0 |
256.498 |
13 |
↓
|
|
|
|
|
|
|
Analogs
-
5065286
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
700 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.47 |
21.02 |
-31.73 |
0 |
1 |
1 |
0 |
388.704 |
19 |
↓
|
|
|
|
Analogs
-
1532339
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYN1-1-E |
Dynamin-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3150 |
0.35 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2100 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
17.81 |
-28.79 |
0 |
1 |
1 |
0 |
312.606 |
17 |
↓
|
|
|
|
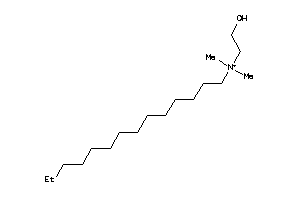
Analogs
-
36177047
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2000 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.77 |
11.27 |
-63.11 |
2 |
4 |
2 |
40 |
374.654 |
19 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
1600 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.33 |
10.79 |
-29.02 |
0 |
1 |
1 |
0 |
186.363 |
8 |
↓
|
|
|
|
Analogs
-
36177047
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
480 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
10.57 |
-28.73 |
1 |
2 |
1 |
20 |
258.47 |
13 |
↓
|
|
|
|
Analogs
-
36177025
-
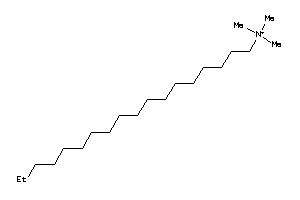
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
700 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
11.58 |
-29.06 |
0 |
1 |
1 |
0 |
200.39 |
9 |
↓
|
|
|
|
Analogs
-
4072653
-
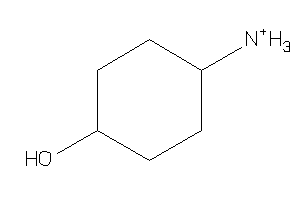
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.52 |
-4.15 |
-40.69 |
5 |
3 |
1 |
62 |
131.199 |
1 |
↓
|
|
|
|
Analogs
-
38433648
-
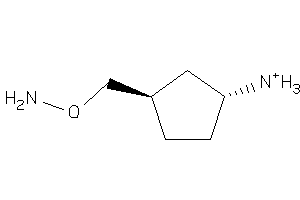
-
38433649
-
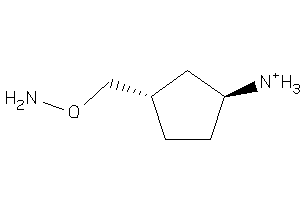
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.41 |
-0.49 |
-44.47 |
5 |
3 |
1 |
63 |
131.199 |
2 |
↓
|
|
|
|
Analogs
-
38433648
-

-
38433649
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.41 |
-3.96 |
-44.44 |
5 |
3 |
1 |
62 |
131.199 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
7943 |
0.79 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
4.52 |
-53.32 |
1 |
2 |
-1 |
17 |
161.275 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.27 |
3.69 |
-41.88 |
1 |
2 |
-1 |
17 |
161.275 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.27 |
3.23 |
-9.47 |
1 |
2 |
0 |
17 |
162.283 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPN-1-E |
Aminopeptidase N (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
22 |
1.34 |
Binding ≤ 10μM
|
|
AMPN-1-E |
Aminopeptidase N (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
22 |
1.34 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
10000 |
0.88 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
-1.82 |
-38.56 |
3 |
1 |
1 |
27 |
134.268 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.15 |
-2.78 |
-33.97 |
3 |
1 |
0 |
27 |
133.26 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
5012 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
12.89 |
-91.14 |
2 |
2 |
2 |
17 |
294.527 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
3100 |
0.64 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
280 |
0.76 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3100 |
0.64 |
Binding ≤ 10μM
|
|
ACHD-2-E |
Acetylcholine Receptor Protein Delta Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.78 |
Binding ≤ 10μM
|
|
ACHA7-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1800 |
0.67 |
Functional ≤ 10μM
|
|
Z104288-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 2), Other |
Other |
3800 |
0.63 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
6310 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
4.59 |
-0.3 |
1 |
1 |
0 |
12 |
167.296 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
220 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.93 |
27.96 |
-76.82 |
0 |
2 |
2 |
0 |
510.98 |
29 |
↓
|
|
|
|
Analogs
-
39268877
-
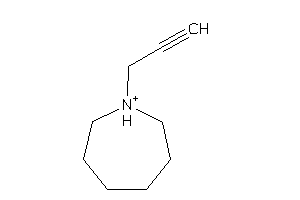
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
360 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
14.07 |
-30.31 |
0 |
1 |
1 |
0 |
252.466 |
12 |
↓
|
|
|
|
|
|
|
Analogs
-
36177025
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
5000 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.18 |
10.02 |
-29 |
0 |
1 |
1 |
0 |
172.336 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
968256
-
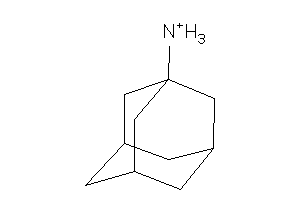
Draw
Identity
99%
90%
80%
70%
Vendors
And 72 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-2-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMD3B-2-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
540 |
0.67 |
Binding ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
9520 |
0.54 |
Binding ≤ 10μM
|
|
Z104297-3-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #3 Of 3), Other |
Other |
5500 |
0.57 |
Binding ≤ 10μM
|
|
Z104298-2-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #2 Of 2), Other |
Other |
5600 |
0.57 |
Binding ≤ 10μM
|
|
Z104297-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2B (cluster #1 Of 1), Other |
Other |
1020 |
0.65 |
Functional ≤ 10μM
|
|
Z104298-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #1 Of 1), Other |
Other |
911 |
0.65 |
Functional ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
1585 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
4.76 |
-42.23 |
3 |
1 |
1 |
28 |
180.315 |
0 |
↓
|
|
|
|
Analogs
-
1532339
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
5.4 |
-28.88 |
0 |
1 |
1 |
0 |
284.552 |
15 |
↓
|
|
|
|
Analogs
-
3871393
-
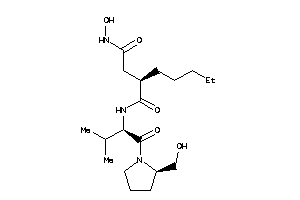
-
3871392
-
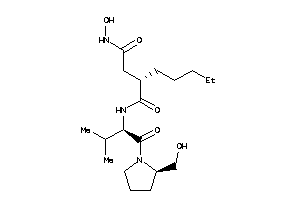
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
5 |
0.43 |
Binding ≤ 10μM
|
|
Q9JN24-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
140 |
0.36 |
Binding ≤ 10μM |
|
DEF1A-1-E |
Peptide Deformylase 1A, Chloroplastic (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.39 |
Binding ≤ 10μM
|
|
DEFM-1-E |
Peptide Deformylase Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.32 |
Binding ≤ 10μM
|
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.28 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.35 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
330 |
0.34 |
Binding ≤ 10μM |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.26 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2512 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
2.32 |
-16.44 |
4 |
8 |
0 |
119 |
385.505 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.51 |
3.4 |
-55.95 |
3 |
8 |
-1 |
122 |
384.497 |
11 |
↓
|
|
|
|
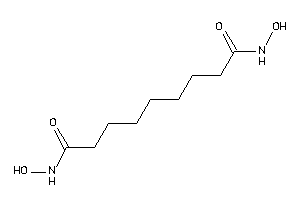
Analogs
-
5178518
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
29 |
0.75 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC5-2-E |
Histone Deacetylase 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.78 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
950 |
0.60 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2239 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
-0.54 |
-22.78 |
4 |
6 |
0 |
99 |
204.226 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.45 |
0.67 |
-63.15 |
3 |
6 |
-1 |
101 |
203.218 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.45 |
1.88 |
-111.59 |
2 |
6 |
-2 |
104 |
202.21 |
7 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
700 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.32 |
21.3 |
-30.07 |
0 |
1 |
1 |
0 |
382.741 |
22 |
↓
|
|
|
|
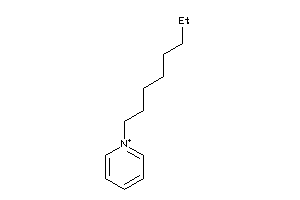
Analogs
-
42784657
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
700 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.03 |
16.54 |
-27.61 |
0 |
1 |
1 |
4 |
304.542 |
15 |
↓
|
|
|
|
Analogs
-
36177047
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
810 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
13.71 |
-28.69 |
1 |
2 |
1 |
20 |
314.578 |
17 |
↓
|
|
|
|
|
|
|
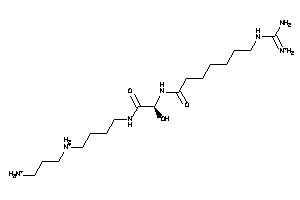
Analogs
-
4215557
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
148 |
0.35 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
81 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.16 |
-1.53 |
-165.74 |
13 |
10 |
3 |
186 |
390.553 |
18 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.16 |
-1.91 |
-89.76 |
12 |
10 |
2 |
185 |
389.545 |
18 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.16 |
-2.88 |
-88.87 |
12 |
10 |
2 |
182 |
389.545 |
18 |
↓
|
|
|
|
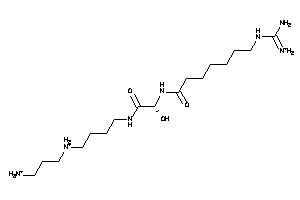
Analogs
-
3775146
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
148 |
0.35 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
81 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.16 |
-2.47 |
-170.31 |
13 |
10 |
3 |
186 |
390.553 |
18 |
↓
|
|
|
|
|
|
|
Analogs
-
5065286
-

-
8214735
-
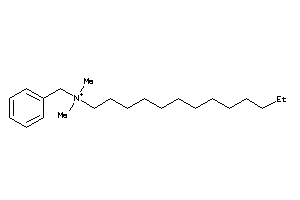
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
1000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
17.89 |
-31.81 |
0 |
1 |
1 |
0 |
332.596 |
15 |
↓
|
|
|
|
|