|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.29 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.33 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.31 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.35 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.34 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
95 |
0.36 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.37 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.37 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.37 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.26 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.27 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.33 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
Q5SC61-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.31 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.40 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.27 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.28 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Functional ≤ 10μM |
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-1-E |
TNF-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
80 |
0.37 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
940 |
0.31 |
Functional ≤ 10μM
|
|
Z80523-1-O |
SW1353 (cluster #1 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
72 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
8.8 |
-21.46 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.25 |
-46.75 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.24 |
-46.38 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.29 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.33 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.31 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
710 |
0.32 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.39 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.35 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.34 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.34 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
M4K4-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.28 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1200 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
95 |
0.36 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.37 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.37 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.38 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.37 |
Binding ≤ 10μM
|
|
MLTK-1-E |
Mixed Lineage Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.28 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.26 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.27 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2700 |
0.29 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.33 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.30 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
Q5SC61-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.31 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.40 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.28 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.27 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.29 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
860 |
0.31 |
Binding ≤ 10μM
|
|
TGFR1-1-E |
TGF-beta Receptor Type I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.27 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.30 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.28 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.29 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.28 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Functional ≤ 10μM |
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-1-E |
TNF-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.37 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
80 |
0.37 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
2500 |
0.29 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
940 |
0.31 |
Functional ≤ 10μM
|
|
Z80523-1-O |
SW1353 (cluster #1 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
72 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
8.81 |
-16.64 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.10 |
8.8 |
-16.98 |
1 |
4 |
0 |
59 |
377.444 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.10 |
9.25 |
-46.22 |
2 |
4 |
1 |
60 |
378.452 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
7 |
0.42 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1500 |
0.30 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.45 |
Binding ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
27 |
0.39 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
45 |
0.38 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.40 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
770 |
0.32 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
2 |
0.45 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
3 |
0.44 |
Functional ≤ 10μM
|
|
Z80409-1-O |
R2 (cluster #1 Of 1), Other |
Other |
7 |
0.42 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
|
Z80459-1-O |
SCC-7 (cluster #1 Of 1), Other |
Other |
6 |
0.43 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
11 |
0.41 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
16 |
0.40 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
5 |
0.43 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.37 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
58 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
5.85 |
-37.21 |
6 |
8 |
1 |
119 |
370.433 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.19 |
5.38 |
-15.09 |
5 |
8 |
0 |
118 |
369.425 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
1000 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
-0.17 |
-4.55 |
1 |
1 |
0 |
15 |
225.316 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
3.03 |
-4.4 |
2 |
8 |
0 |
120 |
322.243 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
36 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
9.45 |
-4 |
2 |
8 |
0 |
121 |
350.297 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.48 |
12.76 |
-11.89 |
1 |
4 |
0 |
51 |
425.507 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.48 |
13.2 |
-34.52 |
2 |
4 |
1 |
52 |
426.515 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.48 |
13.06 |
-51.7 |
2 |
4 |
1 |
52 |
426.515 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.34 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3620 |
0.36 |
Binding ≤ 10μM
|
|
B0BL08-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
10 |
0.53 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2700 |
0.37 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
20 |
0.51 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
300 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
152 |
0.45 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
103 |
0.47 |
Functional ≤ 10μM
|
|
Z50424-1-O |
Cryptosporidium Parvum (cluster #1 Of 2), Other |
Other |
4000 |
0.36 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
9309 |
0.34 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
4.4 |
-13.71 |
4 |
7 |
0 |
106 |
290.323 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.00 |
4.89 |
-42.71 |
5 |
7 |
1 |
107 |
291.331 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACV1B-1-E |
Activin Receptor Type-1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
950 |
0.34 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
AVR2B-1-E |
Activin Receptor Type-2B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4800 |
0.30 |
Binding ≤ 10μM
|
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.35 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.35 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.33 |
Binding ≤ 10μM
|
|
DDR2-1-E |
Discoidin Domain-containing Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.32 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
910 |
0.34 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.32 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.35 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2700 |
0.31 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.32 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
530 |
0.35 |
Binding ≤ 10μM
|
|
KC1AL-1-E |
Casein Kinase I Isoform Alpha-like (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.32 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.40 |
Binding ≤ 10μM
|
|
KC1E-1-E |
Casein Kinase I Epsilon (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.38 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
270 |
0.37 |
Binding ≤ 10μM
|
|
LATS2-1-E |
Serine/threonine-protein Kinase LATS2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.32 |
Binding ≤ 10μM
|
|
MK08-1-E |
Mitogen-activated Protein Kinase 8 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3100 |
0.31 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
51 |
0.41 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.40 |
Binding ≤ 10μM
|
|
MK12-1-E |
MAP Kinase P38 Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.28 |
Binding ≤ 10μM
|
|
MK13-1-E |
MAP Kinase P38 Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.40 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Binding ≤ 10μM
|
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.28 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4200 |
0.30 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.35 |
Binding ≤ 10μM
|
|
NLK-1-E |
Serine/threonine Protein Kinase NLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
|
PTK6-1-E |
Tyrosine-protein Kinase BRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.29 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.32 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.36 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.31 |
Binding ≤ 10μM
|
|
ST32B-1-E |
Serine/threonine-protein Kinase 32B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
STK36-1-E |
Serine/threonine-protein Kinase 36 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.34 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.30 |
Binding ≤ 10μM
|
|
TNIK-1-E |
TRAF2- And NCK-interacting Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.32 |
Binding ≤ 10μM
|
|
TTK-1-E |
Dual Specificity Protein Kinase TTK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.30 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
50 |
0.41 |
Functional ≤ 10μM |
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
50 |
0.41 |
Functional ≤ 10μM |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.29 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
1000 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
8.78 |
-41.1 |
2 |
4 |
0 |
62 |
331.35 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
9.26 |
-92.55 |
3 |
4 |
1 |
63 |
332.358 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.78 |
7.25 |
-9.85 |
2 |
4 |
0 |
62 |
331.35 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
5500 |
0.32 |
Functional ≤ 10μM
|
|
Z50424-1-O |
Cryptosporidium Parvum (cluster #1 Of 2), Other |
Other |
800 |
0.37 |
Functional ≤ 10μM |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
250 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
5.39 |
-10.97 |
2 |
10 |
0 |
155 |
346.365 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1320 |
0.36 |
Binding ≤ 10μM
|
|
Z102116-1-O |
Toxoplasma Gondii RH (cluster #1 Of 2), Other |
Other |
5300 |
0.32 |
Functional ≤ 10μM
|
|
Z50424-1-O |
Cryptosporidium Parvum (cluster #1 Of 2), Other |
Other |
750 |
0.37 |
Functional ≤ 10μM |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2900 |
0.34 |
Functional ≤ 10μM |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
650 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
11.44 |
-4.17 |
0 |
7 |
0 |
95 |
335.282 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.46 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Dihydrofolate Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Functional ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
10 |
0.66 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
80 |
0.58 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
9900 |
0.41 |
Functional ≤ 10μM |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
700 |
0.51 |
Functional ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
5000 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
6.53 |
-28.58 |
5 |
4 |
1 |
79 |
249.725 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.84 |
6.08 |
-5.33 |
4 |
4 |
0 |
78 |
248.717 |
2 |
↓
|
|
|
|
Analogs
-
34016526
-
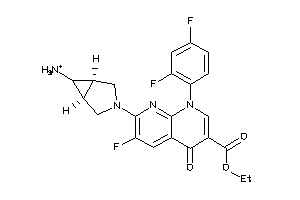
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
3000 |
0.26 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
960 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.80 |
11.67 |
-85.37 |
3 |
7 |
0 |
106 |
416.359 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.80 |
11.62 |
-60.88 |
2 |
7 |
-1 |
104 |
415.351 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
38 |
0.43 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.51 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.45 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
30 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
260 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-3.63 |
-16.02 |
4 |
7 |
0 |
109 |
325.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.41 |
-41.04 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.4 |
-28.87 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|